OBO Foundry Newsletter Issue 9
Over the past two years, this newsletter has reported on community progress, revisions to OBO principles, and developments in shared infrastructure. In this issue, we present updates from the working groups, highlight recent ontology submissions that have been recently accepted or are still undergoing review, and share recent research and tools that strengthen the OBO ecosystem.
The OBO Foundry depends on active community participation. We encourage you to contribute through ontology development, tool building, raising issues, joining discussions, and proposing content for the next newsletter!
OBO Foundry Operations Committee.
Highlights
OBO Dashboard updated
The OBO Foundry Dashboard tracks how well ontologies follow OBO Foundry principles. Ontologies that meet these principles are listed on the OBO Foundry homepage https://obofoundry.org. We encourage ontology developers to check the dashboard regularly to monitor and improve their work.
You can view the most recent dashboard update (last updated 2025-10-07) here: OBO Dashboard (2025-10-07)
Decisions Made and Working Group Updates
The following updates were approved by the Editorial and Technical Working Groups:
- Proper use of the metadata tag “term tracker item” (IAO:0000233): Values should be restricted to URLs/IRIs, not free text. This ensures clear links between terms and their corresponding tracker items.
- New properties in OMO:
- “preferred label for community” (OMO:0002001) is a way to denote a synonym / alternative term for a class (or property) to be preferred by some specific community. For example, in the disease community, different groups prefer to talk about the exact same disease using different labels.
- “has ontology hierarchical property” (OMO:0003014) is a way to document in your own ontology how (based on which properties) you wish hierarchical browsers like OLS and Ontobee to render the hierarchy. Most browsers render rdfs:subClassOf by default, but many ontologies use other properties like “part of”.
- The Editorial Working Group (EWG) is putting together a document that will contain suggestions and recommendations for ontology development for cases deemed too detailed to be a standalone Principle or even to be included as part of a Principles page. The document will include guidelines covering technical considerations, content suggestions, release artefacts, and communication best practices. Thus far, the following guidelines are slated for inclusion:
- Ontology root term annotation
- Proper use of the metadata tag “term tracker item”
Other topics will be added as well (see Ongoing Discussions, below).
Ongoing Discussions
- Minimal ontology-level metadata fields (#1365): Discussion surrounds which metadata fields must or should be included in the ontology header section, such as ontology title, description, license, and others. Particular focus is on the use of dcterms:date for cases were semantic versioning (e.g. v1.1.0) is used.
- Ontology root term annotation (#2149): Allows ontologies to specify ontology-specific root terms, helping browsers display hierarchies more clearly.
- Minimal ontology-level metadata fields (#1365): Two tables will be prepared:
- one with required and recommended fields;
- one with additional potential fields (including those not to use) with guidance.
- Language tag guidance (#479): There is an increasing interest in having OBO ontologies ‘internationalized’ by including, at a minimum, synonyms in other languages. There is an emerging consensus that xsd:string should be omitted for rdfs:label and IAO:0000115. Further discussion is needed on tooling support, default language tags, and consistency across translations.
These items will continue to be revisited in future meetings as progress is made.
Ontologies
New Ontologies Accepted in the OBO Foundry Ontology Library
Exercise Medicine Ontology (EXMO)
- GitHub Issue:#2615
- Key Discussion: EXMO provides standardized terms for exercise physiology, training interventions, performance assessment, and health outcomes related to physical activity. Following review, the ontology was approved for inclusion in the OBO Foundry with consensus and no objections raised.
- See also: Newsletters 8.
New ontologies currently under review
Physiologically Based Pharmacokinetic Modelling Ontology (PBPKO)
- Join the discussion: GitHub Issue: #2563
Key Discussion: PBPKO aims to capture concepts used in physiologically based pharmacokinetic modeling. The ontology has undergone multiple rounds of updates in response to feedback. The most recent submission includes modifications that require reviewer follow-up. The review process remains open until these are assessed. - See also: Newsletter 7.
Experimental Measurements, Purposes, and Treatments Ontology (EMPTY)
- Status: Awaiting updates from submitter
- Join the discussion: GitHub Issue: #2753
- Key Discussion: The ontology proposes to cover experimental measurements, study purposes, and treatment interventions. Initial review identified significant overlap with existing OBO resources. The submitter has requested additional time to revise the scope and address reviewer feedback.
- See also: First introduced in Newsletter 9.
Intrinsically Disordered Proteins Ontology (IDPO)
- Status: Under Review
- Join the discussion: GitHub Issue: #2770
- Key Discussion: The ontology defines concepts and relationships describing the structural states, transitions, and functional roles of intrinsically disordered proteins (IDPs) and regions (IDRs). These proteins lack stable tertiary structures and play key roles in processes such as transcription, translation, signaling, and cell division. The ontology aims to provide a standardized vocabulary to support data integration, annotation, and analysis in structural and molecular biology.
- See also: Newly submitted for review.
A Foundational Ontology for Computational Sociology (OCS)
- Status: Under Review
- Join the discussion: GitHub Issue: #2771
- Key Discussion: The ontology formalizes core sociological concepts and their relationships to support computational social science research. It includes over 700 classes, 250 object properties, and 200 causal relations, all mapped under the Basic Formal Ontology (BFO 2020) framework. OCS enables systematic knowledge representation, automated reasoning about social phenomena, and semantic interoperability across disciplines.
- See also: Newly submitted for review.
Spotlight on well-established OBO ontologies
In this issue, we highlight the Evidence and Conclusion Ontology (eco) from the OBO Foundry family.
Evidence and Conclusion Ontology
Accurate interpretation and reproducibility of scientific conclusions require that every statement or assertion made by a researcher or automated tool be based on supporting evidence that can be shared with, and examined by, the wider scientific community. The Evidence and Conclusion Ontology (ECO) provides terms for the consistent and computable capture of types of evidence used to support scientific statements. ECO began in the early days of the Gene Ontology consortium as a place to contain not only the Gene Ontology evidence codes, but also the wider set of evidence terms in use by the various model organism databases that were part of the consortium. Since then, ECO has grown into an independent ontology used by more than 60 prominent data resources including UniProt and the Alliance of Genome Resources (AGR). Currently ECO contains >2000 active terms encompassing both experimental and computational types of evidence. We continuously add new terms to the ontology in response to requests from our users. Some areas of recent development include terms for machine learning methods developed in collaboration with UniProt, EBI, and AGR as well as terms to describe methods in immune epitope assays in collaboration with the Immune Epitope Database (IEDB). In future work, we will continue to support our existing users, broaden the ECO user community, and integrate ECO with other related ontologies.
Updates on well-established OBO ontologies
Glycosylation Gets a Makeover in the Gene Ontology
The Gene Ontology (GO) recently completed a major revision of its representation of protein glycosylation, particularly N-linked glycosylation (issue #30595). Over time, this branch had accumulated inconsistencies, for example, definitions that included references to arginine and tryptophan N-glycosylation lacking substantial evidence or specific only to bacteria.
The restructuring aligns GO with current biochemical understanding that N-glycosylation primarily occurs via asparagine residues within the Asn-X-Ser/Thr motif, where a preassembled dolichol-linked oligosaccharide is transferred either co- or post-translationally.
The revised hierarchy now distinguishes:
- ER-based glucose/mannose trimming (newly added)
- Golgi apparatus mannose trimming (name updated for clarity)
- N-glycan diversification pathways
- Keratan sulfate proteoglycan biosynthesis (new branch recognizing N-linked vs O-linked variants)
This overhaul involved 8 rounds of discussion and touched 18+ metabolism-related issues. Several of these changes required careful annotation review (go-annotation issues #5827, #5829, #5847, #5859), impacting over 100 experimental annotations across multiple groups. The annotation work involved migrating annotations from obsoleted terms to more specific replacements, reviewing InterPro and Unirule mappings, and ensuring consistency across databases. This demonstrates the collaborative process of modernizing GO to match current glycobiology while maintaining annotation quality.
ChEBI 2.0 redesign
The ChEBI team has launched ChEBI 2.0, a redevelopment of the Chemical Entities of Biological Interest ontology and database. The release updates the entire platform; from infrastructure to user interface; while maintaining data continuity and ontology stability.
Key updates:
- New infrastructure: Rebuilt on modern technologies (PostgreSQL, Django, Vue, Kubernetes) with search powered by Elasticsearch and RDKit.
- Ontology and data products: All downloadable files (ontology, TSVs, SDF, PostgreSQL dump) are now standardized and available at https://ftp.ebi.ac.uk/pub/databases/chebi
- APIs: Legacy SOAP services are deprecated. Users should migrate to the new REST APIs for programmatic access.
- Submission portal: Redesigned and migrated to new backend with improved user management.
- Continuity: All identifiers remain stable and data continue to be released under CC BY 4.0.
Why it matters:
The previous ChEBI codebase had reached its limits. ChEBI 2.0 introduces a simplified schema, faster search, and a scalable foundation for future enhancements; ensuring ChEBI remains a key ontology and data resource for chemical biology.
Milestone:
As of October 2025, the legacy ChEBI website and data releases have been officially retired. Read the full announcement on the ChEMBL blog (See Newsletter 5)
Members and Volunteers
The OBO Foundry is honored to highlight valued community members in each issue. This time, we feature Alexander Diehl.
Alexander Diehl

Alexander Diehl is an Associate Professor at the University at Buffalo. Following training in wet lab molecular biology and immunology and a few years in the biotech industry, Alex joined Mouse Genome Informatics at the Jackson Laboratory in 2003 as a scientific curator and began doing Gene Ontology annotation and development. He has been a project leader of the Cell Ontology since 2005, and has contributed to or developed a number of other biomedical ontologies including the Infectious Disease Ontology, the Ontology for General Medical Sciences, the Neurological Disease Ontology, the Magnetic Resonance Imaging Acquisition and Analysis Ontology, the Cholangiocarcinoma Ontology, and the Ontology for Dental-Care Related Fear and Anxiety. Alex served on the ImmPort and HIPC Data Standards Committee for several years and has served on the OBO Operations Committee since 2018. Alex also teaches undergraduate and graduate courses and mentors students at all levels in Biomedical Informatics at the University at Buffalo, which he joined in 2010.
Spotlight on Research in the OBO community
Expanding and refining the mammalian phenotype ontology to enhance disease model discovery
This recent article highlights major updates to the Mammalian Phenotype (MP) Ontology, an OBO Foundry resource central to disease model discovery. The work focused on expanding disease-relevant terms, improving alignment with the Human Phenotype Ontology (HPO), and strengthening infrastructure for interoperability.
In the past decade the ontology has added over 2,800 new terms including; 140 new terms for pediatric disease models, most notably in congenital heart disease; 124 terms for COVID-19, including diffuse alveolar damage and cytokine storm; and 32 Alzheimer’s-related terms such as amyloid isoform and brain lesion phenotypes. To improve cross-ontology integration, more than 1,500 MP-HPO mappings were created or refined using the SSSOM standard, resolving inconsistencies in labels, definitions, and logical axioms.
Infrastructure and tooling updates further aligned MP with OBO Foundry standards. These included adoption of standardized metadata (ORCID identifiers, date formats), incorporation of uPheno design patterns for logical definitions, and use of ROBOT and the Ontology Development Kit (ODK) for editing, quality control, and release management. Community-facing tools were also strengthened: a new MP-HPO match tool was added to the Human-Mouse Disease Connection (HMDC), and internationalization efforts introduced British spellings and Japanese translations (thanks to the efforts of the MP Japanese Translation Project ).
Together, these developments expand the scope of the MP Ontology and reinforce its alignment with the OBO ecosystem, supporting variant prioritization, disease model discovery, and translational research.
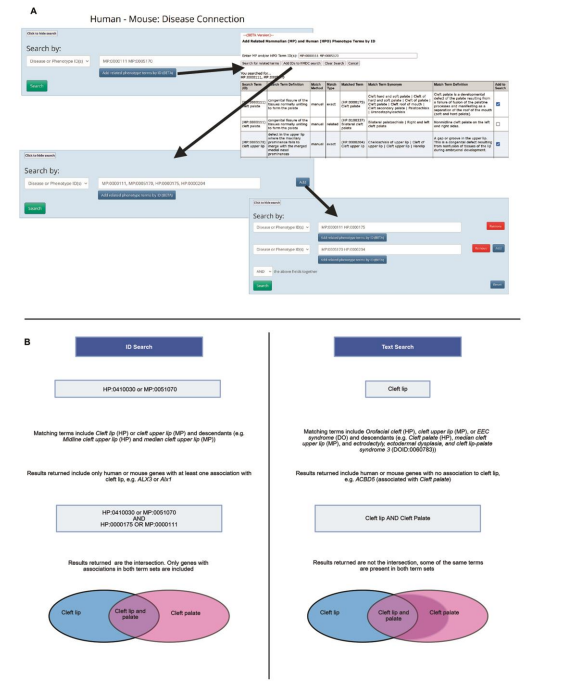
Figure from Susan M. Bello, Anna V. Anagnostopoulos, Leigh C. Carmody, Nicolas Matentzoglu, Cynthia L. Smith. Expanding and refining the mammalian phenotype ontology to enhance disease model discovery. 2025 September 18. https://doi.org/10.1242/dmm.052385
Navigating open-source waters: the pharmaceutical industry’s role in bioontology development
A recent open-access article by OBO Ops member Shawn Tan highlights how the pharmaceutical industry can play a greater role in sustaining OBO ontologies and tools.
This perspective examines how the pharmaceutical industry relies on bioontologies for data integration, FAIR compliance, and AI applications, while development and maintenance remain largely community-driven. The authors identify opportunities for industry to contribute to public efforts, with emphasis on the role of the OBO Foundry and its ecosystem:
- Contributing to ontology content: OBO Foundry ontologies such as Uberon and the Cell Ontology are widely used but resource-constrained. Industry users are encouraged to report issues, propose changes, and contribute pull requests directly to OBO repositories, reducing duplication of in-house fixes and improving interoperability.
- Supporting open-source tooling: OBO Foundry tools such as the Ontology Development Kit (ODK), ROBOT, OAK, and SSSOM provide standardized workflows for ontology management. Active contribution to these tools sustains their development and aligns internal pipelines with community standards.
- Connecting with the community: The OBO Foundry community offers structured entry points (GitHub repositories, OBO Slack, operations committee participation). Engagement enables industry teams to align with ontology development practices, influence standards, and support shared resources such as the Pistoia Alliance’s Pharma General Ontology, which leverages OBO ontologies and LinkML.
Using Novo Nordisk Research & Early Development (R&ED) as a case study, the authors demonstrate how structured engagement with OBO Foundry ontologies and tools can enhance both industry workflows and community resources. They conclude with a call to action for pharma to contribute to the sustainability of open bioontology development.
Mondo: Integrating Disease Terminology Across Communities
The Mondo Disease Ontology is a unified, open-source framework designed to integrate heterogeneous disease terminologies used in research and clinical care. It harmonizes content from 21 disease resources, including OMIM, Orphanet, MeSH, and NCIt, to resolve differences in scope, structure, and naming conventions that limit interoperability and data integration.
Mondo represents approximately 100,000 source concepts as more than 25,000 distinct disease entities encompassing both human and non-human diseases. It includes over 110,000 synonyms, categorized as exact, narrow, broad, or related, following SKOS standards. Each disease entry contains provenance linking definitions and synonyms to original sources.
The ontology supports a pluralistic classification that allows diseases to have multiple parent terms, representing clinical, biological, and genetic perspectives. Logical design templates, Dead Simple OWL Design Patterns (DOSDP), and automated quality control ensure consistent structure and terminology. Mondo’s synchronization workflow imports and aligns updated data from external resources monthly, using a combination of automated matching and manual curation.
Mondo provides curated semantic mappings among major terminologies and distributes these in standardized formats such as SSSOM. Integration with other ontologies, including Uberon, Gene Ontology (GO), and NCBI Taxonomy, links disease terms to anatomical systems, biological processes, and causal organisms. Mondo is used by ClinGen, Open Targets, NORD, OMIM, and MedGen to support consistent disease representation across biomedical databases and clinical applications.
Community contributions are managed through GitHub, where users can propose new terms, report issues, and review changes. More than 5,000 pull requests and 3,900 issues have been recorded since 2017.
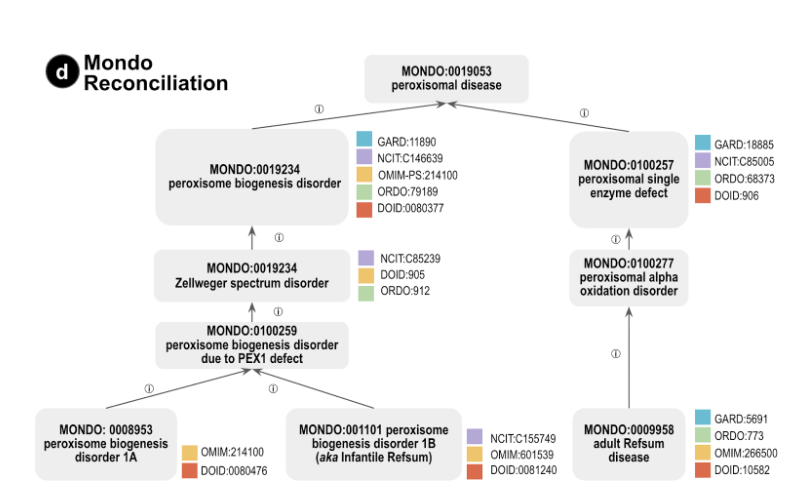
Figure: Portion of Mondo disease hierarchy showing several MYH7-related diseases and whether they are recorded in several major databases. This figure shows how Mondo reconciles these diseases, highlighting subclasses of the term “peroxisomal disease” (MONDO:0019053).
Figure from Nicole A Vasilevsky et al., Mondo: Integrating Disease Terminology Across Communities. Genetics, 2025 October 6. https://doi.org/10.1093/genetics/iyaf215
Chemical classification program synthesis using generative artificial intelligence
This article in Journal of Chemical Informatics introduces the ChEBI Chemical Classifier Program Ontology (C3PO), a novel approach that leverages generative artificial intelligence to create explainable Python programs for chemical structure classification in the ChEBI ontology. Unlike deep learning models that often lack interpretability, C3PO produces deterministic classifiers with natural language explanations for each decision.
The method follows an iterative workflow, Learn–Execute–Iterate–Adapt (LEIA), in which large language models generate candidate classifiers that are refined against benchmark data until they reach acceptable accuracy. Results show strong performance for classes with clear structural patterns, such as glycerol-based lipids, while more complex or ambiguous categories remain challenging. Although deep learning systems like Chebifier outperform C3PO in overall accuracy, the program synthesis approach provides complementary strengths in explainability and curator-guided refinement.
This work highlights how AI-assisted program synthesis can support ontology development and chemical database curation. By combining symbolic, rule-based methods with machine learning, future systems may achieve both transparency and accuracy, advancing sustainable classification in ChEBI and related resources.
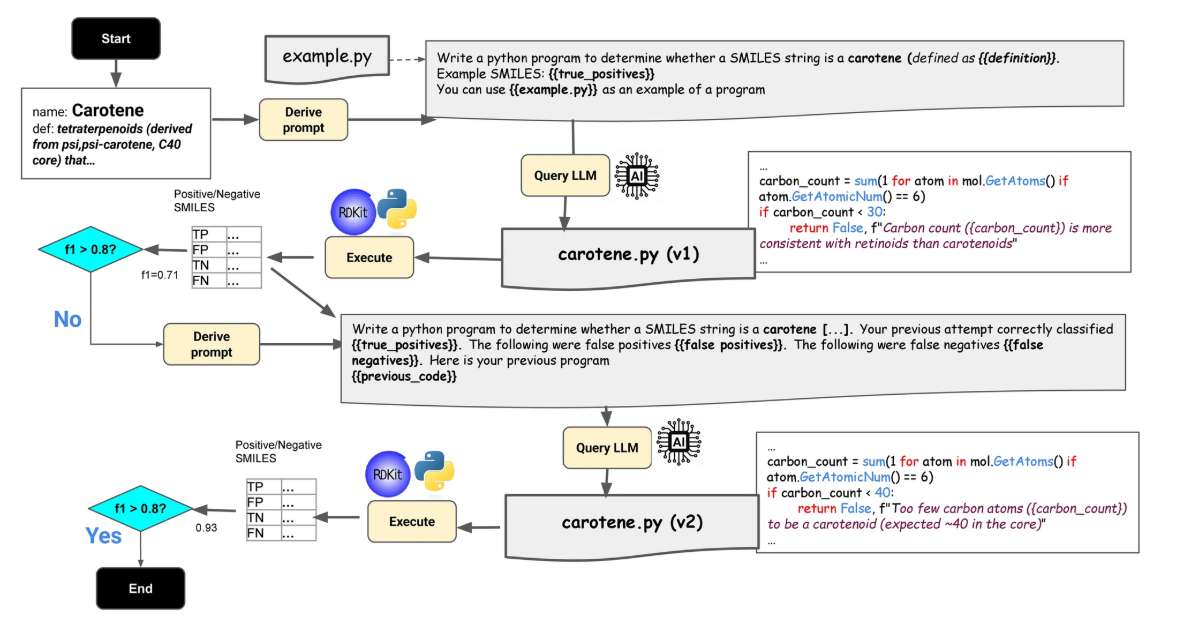
Figure from Christopher J. Mungall, Adnan Malik, Daniel R. Korn, Justin T. Reese, Noel M. O’Boyle & Janna Hastings. Chemical classification program synthesis using generative artificial intelligence. Journal of Cheminformatics volume 17, Article number: 152. 2025 October 1. https://doi.org/10.1186/s13321-025-01092-3
Spotlight on Research: Now Published
Several research papers previously featured in the OBO Newsletter as preprints have now been published in journals.
SeMRA (Semantic Mapping Reasoning and Assembly), “Assembly and reasoning over semantic mappings at scale for biomedical data integration”, featured in issue 8 has been accepted for publication in Bioinformatics (DOI: 10.1093/bioinformatics/btaf542). The paper presents a framework for assembling, validating, and reasoning over large-scale semantic mappings to improve interoperability across biomedical datasets.
DRAGON-AI (Dynamic Retrieval Augmented Generation of Ontologies using Artificial Intelligence), featured in issue 4, now published in Journal of Biomedical Semantics (DOI: 10.1186/s13326-024-00320-3). The paper describes a framework that applies retrieval-augmented generation methods to support the automated creation and refinement of ontologies, integrating AI techniques with ontology engineering workflows.
SPIRES (Structured Prompt Interrogation and Recursive Extraction of Semantics), featured in issue 4, accepted for publication in Bioinformatics (https://doi.org/10.1093/bioinformatics/btae104). The paper presents a method that uses zero-shot learning and structured prompting to extract and organize semantic information for automatically populating knowledge bases.
O3 Guidelines (Open Data, Open Code, and Open Infrastructure) featured in issue 3, now published in Scientific Data (DOI: 10.1038/s41597-024-03406-w). The paper outlines best practices for ensuring sustainable, transparent, and reusable scientific resources through open data, open code, and open infrastructure.
Artificial Intelligence Ontology (AIO) featured in issue 5, now published in Applied Ontology (DOI: 10.1177/15705838241304103). The paper, “The Artificial Intelligence Ontology: LLM-Assisted Construction of AI Concept Hierarchies” presents the LLM-assisted construction of AI concept hierarchies for an ontology of artificial intelligence.
Change Language for Ontologies and Knowledge Graphs featured in issue 6, now published in Database (DOI: 10.1093/database/baae133). The paper, “A change language for ontologies and knowledge graphs”, presents a formal language for representing and managing changes in ontologies and knowledge graphs.
Unified Phenotype Ontology (uPheno) featured in issue 6, now published in Genetics (DOI: 10.1093/genetics/iyaf027). The paper, “The Unified Phenotype Ontology: a framework for cross-species integrative phenomics,” presents a framework for integrating phenotype data across species to support comparative and translational research.
Spotlight on OBO Principles
Principle 7: Relations
To ensure interoperability and logical consistency across OBO Foundry ontologies, existing relations must be reused whenever possible. The Relations Ontology (RO) serves as the primary source for standard relations. By reusing RO terms, ontologies enable automated reasoning, detection of inconsistencies, and inference of new knowledge across domains. When a required relation is not available in RO, developers are expected to search other OBO ontologies before introducing a new one. Relations intended for broader use should be submitted to RO. If a relation is retained within a local ontology, it must be defined as a sub-property of an appropriate RO parent relation, if it exists.
Validation: Each relation that does not use an RO IRI is automatically checked. If a relation label matches one in RO but does not use the correct RO IRI, it is flagged as an error. Any other non-RO relations are reported as information messages. This validation is implemented in the OBO Dashboard to support consistency across ontologies.
Coordination with RO is encouraged through GitHub or the OBO Slack #relation-ontology channel.
For more details, see the OBO Foundry documentation on Principle 7
Spotlight on Tools: Recent updates
Protégé
A new version of the Protégé ontology editor, version 5.6.8, was released in September: https://github.com/protegeproject/protege-distribution/releases/tag/protege-5.6.8
It fixes a bug that, depending on the ontology being edited, could cause frequent complete freezes when browsing the ontology, particularly on MacOS.
Events and News
Registration for the International Conference on Biomedical Ontology (ICBO 2025) is now open:
ICBO 2025 is also accepting submissions for short talks of 10–15 minutes. Submissions should be one-page abstracts and are due by October 24, 2025. Acceptance notifications will be sent on October 30, 2025. These talks are intended to support collaboration with the ICBO community and provide an opportunity for feedback on ongoing work. Abstracts can be submitted through the CMT system: https://cmt3.research.microsoft.com/ICBO2025/Track/6/Submission/Create.
For questions, please contact Asiya Yu Lin (ontology.world@gmail.com).
OBO Academy: Seminar Series (ongoing)
The OBO Academy Seminar Series, organized by members of the Monarch Initiative and open to all, is active again after the summer break. The series offers training and discussions on ontology-related tools and practices. To join, please reach out in the OBO Slack channel #obo-academy.
Upcoming seminars:
- November 11, 2025: Generating SPARQL Queries with ChatGPT (Trish Whetzel)
- December 9, 2025: Efficient Biocuration and Bioinformatics with Claude Code Part 2 (Chris Tabone)
The full schedule and materials are available at https://oboacademy.github.io/obook/courses/monarch-obo-training/
You can also vote for or request new training topics at https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests
Ways to be part of the OBO Foundry community
There are many ways to be part of the OBO Foundry community, beyond using our website to find ontologies of interest. For example:
- Join the obo-discuss mailing list
- If you are interested in the technical aspects, you can also join the obo-tools mailing list
- Join the conversation in our Slack workspace.
- OBO Foundry RSS feed at https://obofoundry.org/feed.xml
- Use our public issue tracker to report a problem you discovered on obofoundry.org or request a new feature
- Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be instead added to the issue tracker for the specific ontology
- Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of our code is publicly readable, you may be able to pinpoint where a bug fix needs to go.
- Request that your ontology be considered for inclusion in the Foundry
- Suggest content for our next newsletter
- If you want to take your contributions to OBO Foundry to the next level, you can nominate yourself to be considered for the OBO Operations Committee. There are many ways in which you can contribute, including assisting with the production of this newsletter.