OBO Foundry Newsletter Issue 8
In this edition, we highlight the recent approval of Principle 19 (Stability of Term Meaning), which sets critical standards for preserving semantic consistency across OBO ontologies. This milestone provides the community with guidance on maintaining the identity of terms and mitigating semantic drift over time. We also provide updates from the OBO Dashboard, recent decisions by the Operations Committee, and progress made by the Editorial and Technical Working Groups.
We report on the continued revision of Principle 7 (Relations), which manages criteria for allocating relation terms to the Relation Ontology (RO) or domain-specific ontologies. Additionally, this issue reviews several ontology submissions that are under active consideration or technical review.
Our spotlight series features the Relation Ontology (RO) and Unified Phenotype Ontology (uPheno), alongside community-contributed research on semantic interoperability and cross-species phenotype integration.
As always, we encourage you to participate in the OBO community, whether through contributions, feedback, or development efforts.
OBO Foundry Operations Committee.
Highlights
OBO Dashboard updated
The OBO Foundry Dashboard helps track the progress of ontologies in aligning with OBO Foundry principles. Ontologies that meet these standards are featured on the OBO Foundry homepage at https://obofoundry.org. We encourage all ontology developers to regularly consult the dashboard to assess and enhance their ontologies.
You can view the most recent dashboard update (last updated 2025-05-29) here: OBO Dashboard
Decisions Made and Working Group Updates
This period saw continued progress by the Editorial and Technical Working Groups, alongside several key decisions by the OBO Operations Committee. The Editorial Working Group (EWG) received formal approval and publication of Principle 19 (Stability of Term Meaning), which sets forth best practices for versioning and maintaining consistent ontology releases. The EWG also improved Principle 12 (Naming Conventions) by clarifying when term corrections can occur without triggering obsoletion, promoting more consistent terminology across OBO ontologies.
Ongoing Discussions
The Editorial Working Group is currently revising Principle 7 (Relations), which covers the placement of relationship terms across domain-specific ontologies and the Relation Ontology (RO). Committee members are debating criteria such as whether both the domain and range of a relation must exist in the same ontology, and whether logical invertibility is a practical requirement. The goal is to provide clearer guidance for ontology developers while maintaining flexibility across disciplines. A draft revision is in progress, but no consensus has yet been reached. Join the discussion
Ontologies
New Ontologies Accepted in the OBO Foundry Ontology Library
- GitHub Issue: #2604
- Key Discussion: The Biomarker Ontology provides terms relevant to identifying and describing biological markers in health and disease contexts. The submitter engaged in active feedback resolution and coordinated with the developers of the Ontology of Biological and Clinical Investigations (OBCI) to align future relation modeling. A key point of discussion was whether approval could be granted based on the commitment to update relations once OBCI terms are integrated into the Relations Ontology (RO). The committee agreed that this commitment was sufficient, and the ontology was approved after a public comment period with no objections.
New ontologies currently under review
Physiologically Based Pharmacokinetic Modelling Ontology (PBPKO)
- Status: Reviewer response required
- Join the discussion: GitHub Issue: #2563
- Key Discussion: See Newsletter 7.
Radiation Therapy Ontology (RTO)
- Status: Reviewer assigned; domain expert still needed
- Join the discussion: GitHub Issue: #2683
- Key Discussion: The Radiation Therapy Ontology (RTO) provides standardized terms for radiation procedures, equipment, dosages, and treatment planning. While it passed most of the technical review, it did not comply with Principle 3 due to the use of non-OBO-compliant IRIs. The committee clarified that a valid namespace must be in place at submission. A reviewer has been assigned, but a domain expert is still needed to complete the evaluation.
Experimental Model Ontology (EXMO)
- Status: Resubmitted; in technical review
- Join the discussion: GitHub Issue: #2615
- Key Discussion: The Exercise Medicine Ontology (EXMO) aims to support personalized exercise prescriptions by standardizing terms related to physical activity, health status, and exercise interventions. While recent updates corrected identifier formatting and addressed several reviewer concerns, issues remain with reuse of out-of-scope terms that should be imported from existing OBO ontologies, such as CMO and DOID. Additional revisions are needed to improve modeling of clinical measurements, consistency in axioms for exercise equipment, and completeness of term definitions. The reviewers acknowledged substantial progress and requested the submitter document future term requests to external ontologies. The review remains open while these refinements are implemented.
Spotlight on well-established OBO ontologies
In this issue, we highlight two ontologies from the OBO Foundry family, the Relation Ontology (RO) and the Unified Phenotype Ontology (uPheno).
The Relation Ontology (RO) was designed to classify and organize relationships between entities in the life sciences and biomedicine. Each RO relation is uniquely identified by a Uniform Resource Identifier (URI) and comes with additional information like a human-readable label, synonyms, and definitions, which can be reused in ontologies and databases. RO includes both general-purpose relations, such as part of, and more specific relations applicable to particular domains. Its core relations (“RO Core”) are defined using the domain-independent from the Basic Formal Ontology (BFO) or the Core Ontology of Biology and Biomedicine (COB). RO provides precise semantics for its relations, defining features such as Transitivity, Symmetry, and Inverses. This allows the use of deductive reasoners to infer new relationships and detect inconsistencies. RO is enshrined as the standard ontology for relationship types in the OBO community. The reuse of RO relations is important for maintaining ontology interoperability. Currently, over 140 OBO Foundry ontologies use RO. RO is used across diverse biological and biomedical domains, including neuroscience, mechanistic modeling, environmental sciences, anatomy and development, phenotypes and diseases, and biomedical investigations. It is also used in various databases and knowledge graphs, such as the Virtual Fly Brain. Beyond the OBO ecosystem, RO properties are used in Wikidata, where there are currently 20 Wikidata properties linked to RO properties via the property “Relations Ontology ID” (as of 2021). These properties are used in 13,167,119 statements with ‘in taxon,’ ‘part of,’ ‘located in,’ and ‘has part’ used more than a million times each. The Biolink Model also uses predicates mapped to RO. RO is publicly available on GitHub https://github.com/oborel/obo-relations. Monthly editors meetings are open to the community, more information can be found here.
The Unified Phenotype Ontology (uPheno) brings together various species-specific phenotype ontologies to enable cross-species analysis of phenotype data. The twelve participating ontologies span from vertebrates to single-cell organisms. The focus so far has been on phenotypic changes induced by genetic and environmental manipulations of organisms (Fig. 1), as this is the focus of the participating ontologies. To facilitate the integration of these ontologies, the uPheno working group has developed a library of patterns that can be used to generate standardized terms, including term labels, synonyms, and text and logical definitions. The framework also helps bootstrap entirely new ontologies (e.g., XPO, PLANP, ZP). These patterns have been adopted by the member ontologies to create new terms and/or manage logical definitions for existing terms. At the Biocuration conference in April, we held a workshop to discuss how to improve integration and enhance the upper-level structure of uPheno to increase usability. uPheno currently integrates 12 species-specific ontologies and offers SSSOM-formatted cross-species mappings (e.g., between HPO and MP). This supports critical use cases such as disease gene discovery and variant prioritization, powering tools like Exomiser and the Monarch Initiative.
The ontology files and patterns are all available on the uPheno GitHub site (https://github.com/obophenotype/upheno). The working group meets monthly to discuss issues, please contact info@monarchinitiative.org to receive an invitation. In addition, there is a #upheno channel in the OBO-Community Slack and a dedicated uPheno Slack workspace.
Read more about uPheno in the 2025 Genetics publication: https://doi.org/10.1093/genetics/iyaf027.
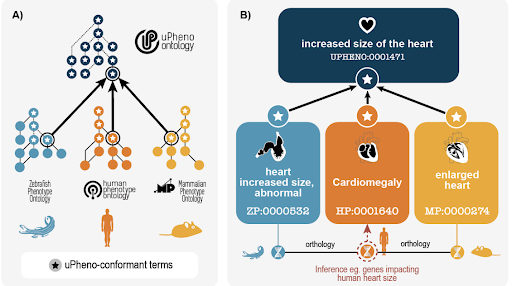
Figure 1. uPheno brings together species-specific ontologies within a common framework to facilitate translational research (from https://doi.org/10.1093/genetics/iyaf027).
Members and Volunteers
The OBO Foundry is honored to highlight two valued community members.
Lynn Schriml

As PI of the Disease Ontology project (http://www.disease-ontology.org/) a NIH/NHGRI-funded Genomic Resource and Knowledgebase, established in 2003, Lynn has been active in the OBO Foundry since 2008. She currently contributes to the OBO Operations team, reviewing ontologies, representing the OBO Foundry at conferences, and has previously worked on the OBO Editorial Working Group.
As an Associate Professor at the University of Maryland School of Medicine, her research includes the classification of standardized descriptors of biomedical concepts. She has been involved with the development of a suite of OBO Foundry biomedical ontologies including the Human Disease Ontology, Symptom Ontology, Influenza Ontology, Pathogen Transmission Ontology, Disease Drivers Ontology, and has contributed to the GAZ geographic location gazetteer and the ENVO environment ontology.
A key component of her ontology development has involved ongoing engagement with disease-specific research communities and ontology developers (e.g., Model Organism Databases – drosophila, zebrafish, mouse, rat, yeast and worm, Human Phenotype Ontology, Mammalian Phenotype Ontology, Alliance of Genome Resources) to include extension, revisions and additions to the Disease Ontology based on disease models defined by the research community.
Bjoern Peters

Dr. Bjoern Peters is a Professor at the La Jolla Institute for Immunology in San Diego, California. Since starting his Ph.D. in theoretical biophysics at Humboldt University in Berlin, Germany, he has worked on the development and validation of machine learning tools to analyze and predict which parts of a pathogen, allergen, or cancer cell are targeted by immune responses. Identifying these specific molecular targets of immune responses, called epitopes, recognized by diseased individuals opens a path toward developing diagnostics, vaccines, and therapeutics.
In 2004, he moved to the La Jolla Institute as a postdoc and later became the bioinformatics lead of the Immune Epitope Database (IEDB; PI: Alessandro Sette), which catalogs epitope recognition data curated from the literature. The desire to capture data for the IEDB in a compatible fashion with other databases brought him into contact with the OBO community, and he teamed up with multiple other researchers working on standardizing data capture from a broad range of experimental techniques. This led to the founding of the Ontology of Biomedical Investigations (OBI) in 2006, which he continues to be heavily involved in.
Beyond OBI, Dr. Peters has contributed to many OBO ontologies, the OBO Foundry, and its transition to an ‘operations-driven-project’. He co-led the OBO-services grant with Dr. Chris Mungall and co-initiated the COB project that aims to develop an integration ontology layer for OBO ontologies below BFO. Beyond the IEDB, Dr. Peters has utilized the OBO data standardization techniques for multiple other public-facing database projects as well as for data capture and analysis for data generated in the experimental part of his lab.
Spotlight on Research in the OBO community
Suggestions for extending the FAIR Principles based on a linguistic perspective on semantic interoperability
This article explores the FAIR Guiding Principles emphasize making data Findable, Accessible, Interoperable, and Reusable, yet they do not extensively define the concept of semantic interoperability. In a recent publication, Vogt et al. examine this gap from a linguistic perspective, underscoring the importance of shared understanding for both humans and machines to interpret data effectively.
The authors introduce an enhanced model of semantic interoperability that differentiates between terminological and propositional interoperability, analogous to how language users comprehend words and sentences. They contend that merely aligning terminologies is insufficient; instead, data should be structured like language, with consistent syntax, semantics, and common background knowledge to facilitate reliable interpretation and utilization by humans and machines.
To advance the FAIR framework, they propose enhancements, which they refer to as progressing toward FAIR 2.0, such as developing a comprehensive FAIR ecosystem comprising services for terminology management, schemas, and operational procedures inspired by linguistic models that support meaningful communication.
This interdisciplinary research connects ontologies, linguistics, and data science, highlighting that achieving true interoperability requires establishing shared linguistic and digital understanding.
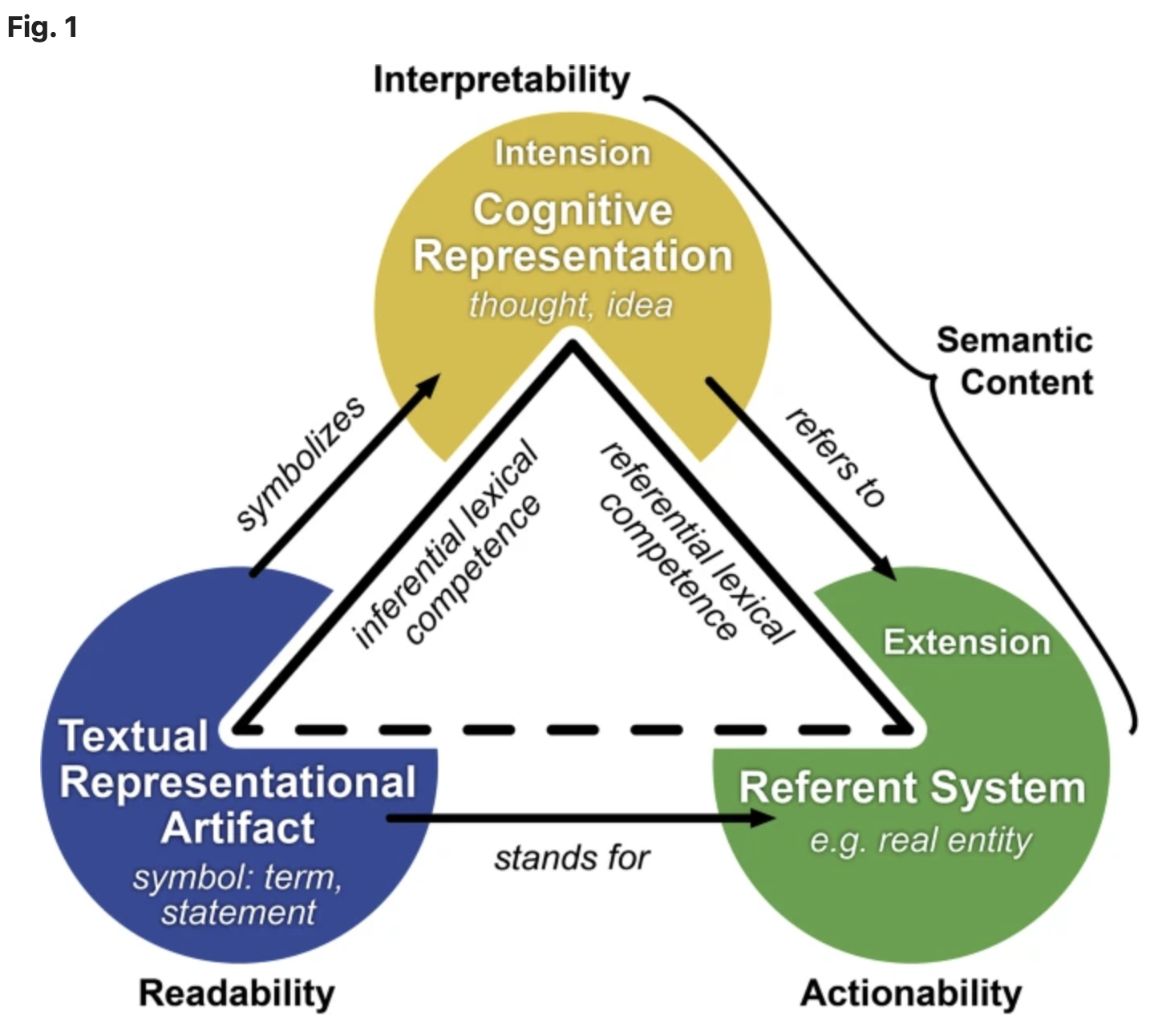
Figure from Lars Vogt, Philip Strömert, Nicolas Matentzoglu, Naouel Karam, Marcel Konrad, Manuel Prinz, Roman Baum. Suggestions for extending the FAIR Principles based on a linguistic perspective on semantic interoperability. 2025 April 24; https://doi.org/10.1038/s41597-025-05011-x
BioPortal: an open community resource for sharing, searching, and utilizing biomedical ontologies
BioPortal is not new (it was first released in 2008), but a new publication highlights recent additions to its functionality. BioPortal (https://bioportal.bioontology.org), developed by the National Center for Biomedical Ontology (NCBO), is the world’s most comprehensive repository of biomedical ontologies. It aims to disseminate biomedical knowledge through principled ontologies, ensuring semantic interoperability across the research landscape.
BioPortal automatically indexes all OBO Foundry ontologies (among others), monitoring them daily and reloading them upon detecting updates. While the OBO Foundry serves as a registry, BioPortal enhances usability by offering a browsable interface and downloadable ontology files (where licensing permits). As of March 2025, BioPortal hosts 1,549 ontologies (1,182 public), representing over 15 million classes and more than 101 million cross-ontology mappings. Its services support a vibrant community, with 18,000+ registered users and over 159 million API calls monthly.
Key features for OBO users include:
- Ontology Browsing and Search: Advanced search tools and collection filters allow intuitive access to OBO and other biomedical ontologies.
- Detailed Ontology Summaries: Each ontology has a dedicated page with metadata, version history, usage metrics, reusable views, and associated projects.
- Class Exploration: Hierarchical class browsing with improved usability.
- Cross-Ontology Mappings: Automatically generated mappings visualize how terms are used across ontologies.
- Annotation Services: The Annotator and Annotator+ services identify ontology terms in text, with support for negation detection.
- REST API: Extensive programmatic access enables integration with external systems like EHRs, decision support tools, and text mining platforms.
- Knowledge Graph Support: KG-BioPortal uses the Biolink Model to merge and query ontologies as a unified knowledge graph.
A standout capability is BioPortal’s interface for suggesting ontology edits. Using the Knowledge Graph Change Language (KGCL), user-submitted changes are converted into GitHub issues, streamlining curator workflows.
BioPortal’s infrastructure supports the broader OntoPortal Alliance, with additional portals serving domains like agriculture and ecology. Looking ahead, BioPortal plans to enhance knowledge graph features, improve metadata (FAIR-aligned), expand mapping support (including SSSOM), and integrate LLMs for smarter annotation.

Figure from Jennifer Vendetti , Nomi L Harris , Michael V Dorf , Alex Skrenchuk , J Harry Caufield , Rafael S Gonçalves , John B Graybeal , Harshad Hegde , Timothy Redmond , Christopher J Mungall , Mark A Musen. BioPortal: an open community resource for sharing, searching, and utilizing biomedical ontologies. 2025 May 13; https://doi.org/10.1093/nar/gkaf402
Assembly and reasoning over semantic mappings at scale for biomedical data integration
A challenge in the automated integration of biomedical data and knowledge is the heterogeneous usage of ontologies and databases that assign their own identifiers to the same or related concepts. Resolving these overlaps requires high availability and coverage of semantic mappings. However, available mappings are incomplete and fragmented across individual resources, motivating their large-scale integration and processing. Therefore, the authors developed the Semantic Mapping Reasoning Assembler (SeMRA), a software tool with the following components:
- A high performance object model for semantic mappings (based on SSSOM)
- A provenance model for automatically generated mappings
- A confidence model granular at the curator-level, mapping set-level, and community feedback-level
- Functionality for assembling, reasoning over, and inferring semantic mappings at scale
SeMRA represents mappings as a directed graph and applies inference algorithms such as inversion, mutation, and transitivity to infer new connections between concepts while tracking provenance and confidence. Its modular design enables import from standard formats (e.g., OBO, OWL), custom sources (e.g., OMIM, ChEMBL), and additional plugins, with all references validated against the Bioregistry. SeMRA includes a web application for interactive exploration, quality control, and curation, and supports export in SSSOM and Neo4j formats. The authors demonstrated SeMRA first by aggregating 43.4 million mappings from 127 sources that jointly cover identifiers from 445 ontologies and databases that they made available on Zenodo (https://doi.org/10.5281/zenodo.11082038). Second, they also demonstrated how SeMRA can be configured with a declarative specification and create five domain-specific mapping sets for anatomical terms, cells/cell lines, genes, protein complexes, and diseases (see https://github.com/biopragmatics/semra/tree/main/notebooks/landscape). Their meta-analysis showed that the integration and inference of domain-specific mappings resulted in meaningful consolidation of redundant terms, especially for the disease and protein complexes. Notably, they reproduced and extended the analysis on the disease landscape presented by Haendel et al., 2020 in “How many rare diseases are there?” (https://doi.org/10.1038/d41573-019-00180-y) to incorporate more disease resources.
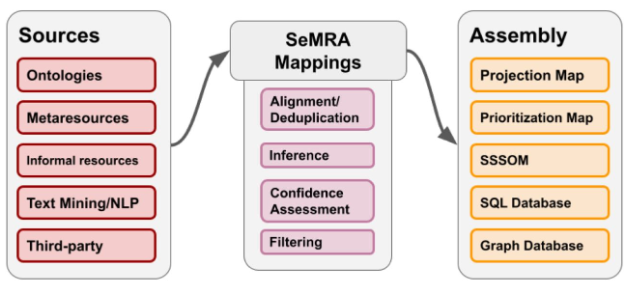
Figure from Charles Tapley Hoyt, Klas Karis, Benjamin M. Gyori. Assembly and reasoning over semantic mappings at scale for biomedical data integration. 2025 April 16; https://www.biorxiv.org/content/10.1101/2025.04.16.649126v1.full
The Cell Ontology in the age of single-cell omics (PREPRINT)
Single-cell omics technologies have transformed our understanding of cellular diversity by enabling high-resolution profiling of individual cells. However, the unprecedented scale and heterogeneity of these datasets demand robust frameworks for data integration and annotation. The Cell Ontology (CL) has emerged as a pivotal resource for achieving FAIR (Findable, Accessible, Interoperable, and Reusable) data principles by providing standardized, species-agnostic terms for canonical cell types - forming a core component of a wide range of platforms and tools. In this paper, we describe the wide variety of uses of CL in these platforms and tools and detail ongoing work to improve and extend CL content including the addition of transcriptomically defined types, working closely with major atlasing efforts including the Human Cell Atlas and the Brain Initiative Cell Atlas Network to support their needs. We cover the challenges and future plans for harmonising classical and transcriptomic cell type definitions, integrating markers and using Large Language Models (LLMs) to improve content and efficiency of CL workflows.
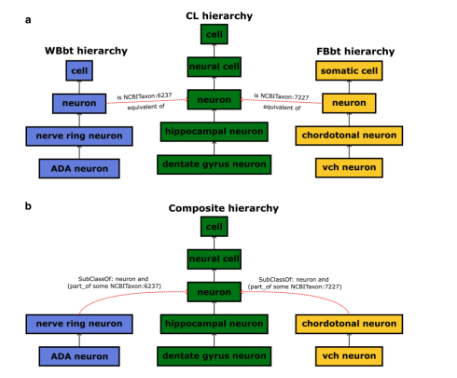
Figure from Shawn Zheng Kai Tan, Aleix Puig-Barbe, Damien Goutte-Gattat, Caroline Eastwood, Brian Aevermann, Alida Avola, James P Balhoff, Ismail Ugur Bayindir, Jasmine Belfiore, Anita Reane Caron, David S Fischer, Nancy George, Benjamin M Gyori, Melissa A Haendel, Charles Tapley Hoyt, Huseyin Kir, Tiago Lubiana, Nicolas Matentzoglu, James A Overton, Beverly Peng, Bjoern Peters, Ellen M Quardokus, Patrick L Ray, Paola Roncaglia, Andrea D Rivera, Ray Stefancsik, Wei Kheng Teh, Sabrina Toro, Nicole Vasilevsky, Chuan Xu, Yun Zhang, Richard H Scheuermann, Chirstopher J Mungall, Alexander D Diehl, David Osumi-Sutherland. The Cell Ontology in the age of single-cell omics. 2025 June 10; https://arxiv.org/abs/2506.10037
Spotlight on OBO Principles
Principle 19: Stability of Term Meaning
Principle 19 of the OBO Foundry emphasizes that the meaning of a term must remain stable over time. Specifically, the *referents *the real-world entities a term denotes must not change as a result of edits to its definition or logical axioms. If a proposed change would shift what the term refers to, a new term with a new IRI and definition must be created instead. This helps protect users from semantic drift and ensures that both humans and machines can rely on the identity and consistency of ontology terms.
The principle allows minor edits for grammar or clarity, as long as these do not alter the referent. It also outlines best practices for handling term obsoletion when meanings change significantly, including when definitions become too vague, inaccurate, or misaligned with scientific understanding. Developers are encouraged to provide replacement guidance and consider the impact on downstream users.
For implementation, the principle specifies exact steps for obsoleting terms in OWL and OBO formats, including use of owl:deprecated, replacing logical axioms, and adding structured metadata like replaced_by or consider. All obsolete term labels must be prefixed with “obsolete ” (lowercase and space included) to comply with Principle 12 (Naming).
The adoption of Principle 19 marks an important milestone for the OBO Foundry. It provides a robust standard for semantic versioning and long-term reliability of ontologies. We encourage all ontology maintainers to read the full principle and begin applying its guidance in their workflows to strengthen trust and usability across the community.
Spotlight on Tools: Recent updates
ROBOT v1.9.8
The ROBOT ontology toolset (v1.9.8), released recently, introduces functional improvements relevant to OBO Foundry ontology developers. The update includes new features, library upgrades, and a series of bug fixes.
New functionality includes:
- A –clean-obo option for the convert command, supporting format cleanup.
- Interpolation of ontology and version IRIs within annotation values.
- Support for selecting punned entities using the remove and filter commands.
- Extensions to the template system allowing predicate-object pair specification.
Library and command updates:
- Apache POI upgraded for improved Excel processing.
- Newline-separated value handling added via SPLIT=\n.
- The reduce command now retains SubObjectPropertyOf axioms.
Bug fixes address:
- Output stability and memory use in owl-diff.
- Exclusion of deprecated terms in lowercase_definition checks.
- Improved handling of axiom annotations during merges.
- Clarified template behavior regarding non-ROBOT columns.
- Resolution of subproperty redundancy detection (issues #1014, #1208).
- Support for embedded default queries in JARs.
- Correction of merging behavior for annotated and unannotated axioms.
The release is available on GitHub and includes three downloadable assets.
Feedback and bug reports: https://github.com/ontodev/robot/issues
Ontology Development Kit (ODK) v1.6:
The ODK is a toolbox and workflow system to manage the ontology lifecycle. After more than a year of work since the last major release, version 1.6 has been published on the 29th May.
This version of the ODK adds major new capabilities, including:
- A redesigned update_repo command for easier repository upgrades
- A new ROBOT plugin with commands for efficient subsetting and upper ontology alignment checking.
- Significant improvements in user management in the interplay with docker.
- Many improvements in the executable workflows (“Makefile”), making them faster and more resilient.
- Ability for SQLite/SemSQL format export, for use with tools like the Ontology Access Kit.
- A new test checks the validity of ID policy files, and another ensures ontologies are aligned with upper-level ontologies when configured.
- Better support for managing SSSOM mapping sets, including merging and selective publishing.
- Tooling improvements including new utilities like jinjanate (replacing j2cli), dicer-cli for ID range management, scala-cli (replacing Ammonite), and yq-mf for YAML/JSON/XML processing. Updates to ROBOT, SSSOM, KGCL, LinkML, and other key packages round out this feature-packed release.
Feedback and bug reports: https://github.com/INCATools/ontology-development-kit/issues
curies v0.10.13:
curies is a python based package for managing IRI-curie conversions. An extension to the curies.Converter class that can apply pre-and post-process rules to CURIEs and URIs was added in v0.10.13. For more information about Curies see Newsletter issue 2. A tutorial is available at https://curies.readthedocs.io/en/latest/preprocessing.html
- An extension to the curies.Converter class that can apply pre-and post-process rules to CURIEs and URIs was added in v0.10.13. A tutorial is available at https://curies.readthedocs.io/en/latest/preprocessing.html
import curies
from curies import PreprocessingRules, PreprocessingConverter, PreprocessingRewrites
rules = PreprocessingRules(
rewrites=PreprocessingRewrites(
full={"OBO_REL:is_a": "rdfs:subClassOf"},
),
)
converter = curies.get_obo_converter()
converter = PreprocessingConverter.from_converter(
converter, rules=rules,
)
>>> converter.parse_curie("OBO_REL:is_a")
ReferenceTuple('rdfs', 'subClassOf')
- A hook for standardizing local unique identifiers was added in v0.10.8. This enables implementing a
curies.Converterthat validates LUIDs with regular expressions, e.g., from the Bioregistry and also that removes redundant prefixes (i.e., bananas) - CURIE parsing in
curies.Converter.parse_curiewas re-implemented to take into account the prefix map, rather than just splitting on a colon : in v0.10.7 - A new function
curies.Converter.parsethat can handle both CURIEs or URIs was added in v0.10.7
Feedback and bug reports: https://github.com/biopragmatics/curies/issues
Spotlight on LinkML
Bridging the Gap for Semantic Data Modeling
The Linked Data Modeling Language (LinkML) is increasingly utilized to support structured data modeling and enhance interoperability across ontologies and related data systems.
Ontologies provide a structured vocabulary for describing scientific concepts. However, researchers often collect data in formats like spreadsheets, where the meaning is only implied. Column headers, value types, and expected formats are rarely standardized, making it difficult and error-prone to integrate datasets across different laboratories or disciplines.
LinkML addresses this challenge by offering a schema framework that makes these underlying assumptions explicit and computable. It allows researchers to define classes, constrain values using ontologies such as ENVO or Uberon, and clearly document expectations about data structure, units, and formats, all in a way that is understandable for both humans and machines.
In practice, LinkML helps bridge the gap between domain knowledge (ontologies) and data structure (schemas), making the validation, integration, and reuse of datasets easier. LinkML not only supports ontologies; it also enhances their utility in real-world data workflows, helping to make scientific data FAIR (Findable, Accessible, Interoperable, and Reusable) and ready for reuse.
Learn more and get started:
Events and News
OBO at Biocuration 2025
The OBO Foundry community participated in the Biocuration 2025 conference at the Stowers Research Institute in Kansas City, Missouri.
Collaborative Biocuration and Training**
**A workshop on collaborative biocuration, co-led by Sabrina Toro (Monarch Initiative) and Nicole Vasilevsky (Critical Path Institute), presented approaches from the OBO Foundry to strengthen and support collaborative work and OBO Academy for providing accessible, version-controlled training resources. The session included case studies from PomBase, the Gene Ontology Consortium, and WormBase’s AI-supported annotation. Key topics were transparency, participation, and quality control in curation. Participants suggested developing a centralized training hub, broader certification options, and increased community exchange.
Slides are available here. Workshop summary is available here.
uPheno workshop: Enhancing the Unified Phenotype Ontology to Support Cross-Species Phenotype Interoperability
Led by Sue Bello (MGI) and Arwa Ibrahim (Monarch Initiative, EBI), the 4th uPheno workshop engaged participants from various ontologies and databases, including the plant community, the Rat Genome Database (RGD), the Mammalian Phenotype Ontology (MP), the Zebrafish Information Network (ZFIN), and the Database Center for Life Science (DBCLS), among others. The workshop focused on two main topics: 1) Improving the upper-level structure of the ontology and 2) Identifying integration and organization issues. Discussions also explored the current challenges in browsing the ontology, uPheno’s classification philosophy, reference ontology dependency, the utility of patternization for complex phenotypes, improving term labels and human-centric definitions, as well as leveraging AI to identify classification discrepancies. Future plans include working with plant ontology curators to integrate plant terms and developing collaborations with the RGD. Participants were encouraged to join the uPheno slack community, attend uPheno monthly calls, and create tickets in uPheno’s GitHub repository.
Slides are available here.
Ontology Automated Workflow for Ontology Community Curation
As part of her podium presentation about the Mondo disease ontology’s community approach to ontology maintenance, Sabrina Toro (Monarch Initiative) presented a workflow that allows the automated import of expertly curated community content directly into the ontology. This workflow relies on a simple spreadsheet maintained at the community source where the curation takes place. The content of this spreadsheet is automatically imported, quality-controlled, and added to the ontology with full provenance and attribution to the community source. This workflow is scalable and has been successfully implemented, for example, to maintain the Mondo rare disease subset.
Slides are available here
AI-Assisted Curation Tools
Presented by Harry Caufield, this podium presentation introduced the open-access tools OntoGPT and CurateGPT, which leverage large language models alongside trusted knowledge bases and literature sources. These tools enable biocurators to efficiently extract, annotate, and curate information from diverse scientific texts, linking extracted concepts directly to ontologies and supporting literature. By streamlining the curation process and improving data provenance, OntoGPT and CurateGPT help curators keep pace with the rapidly growing volume of scientific data.
Slides are available here
Overall, the OBO Foundry’s involvement at Biocuration 2025 focused on training, ontology integration, curation workflows, and the intersection of AI and biocuration.
OBO Academy: Seminar Series (ongoing)
- The OBO Academy: Seminar Series is run by members of the Monarch Initiative, but open to all. Please reach out in the OBO slack channel “#obo-academy” to be signed up.
- Schedule: https://oboacademy.github.io/obook/courses/monarch-obo-training/
- You can vote for and request training topics here: https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests
Ways to be part of the OBO Foundry community
There are many ways to be part of the OBO Foundry community, beyond using our website to find ontologies of interest. For example:
- Join the obo-discuss mailing list
- If you are interested in the technical aspects, you can also join the obo-tools mailing list
- Join the conversation in our Slack workspace.
- OBO Foundry RSS feed at https://obofoundry.org/feed.xml
- Use our public issue tracker to report a problem you discovered on obofoundry.org or request a new feature
- Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be added to the issue tracker for the specific ontology
- Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of our code is publicly readable, you may be able to pinpoint where a bug fix needs to go.
- Request that your ontology be considered for inclusion in the Foundry
- If you want to take your contributions to OBO Foundry to the next level, you can nominate yourself to be considered for the OBO Operations Committee. There are many ways in which you can contribute, including assisting with the production of this newsletter.