OBO Foundry Newsletter Issue 5
Welcome to the 5th issue of the OBO Foundry Newsletter. In this edition, we highlight the progress and achievements of our ontology developers and researchers in driving innovation and collaboration in biomedical research.
The biggest news in this issue is the change to the OBO Foundry homepage to highlight ontologies that meet OBO Foundry principles (as determined by the OBO Dashboard). We also discuss the ongoing efforts to enhance metadata standards, the decision about new ontology submissions, and the various initiatives undertaken by the OBO Operations Committee to streamline processes and foster active participation.
This issue also highlights well-established ontologies, dedicated members and volunteers, advanced research, foundational principles, and essential tools. We provide updates on past events and announce the numerous upcoming opportunities for learning and collaboration, which promise to shape the future of biomedical research.
Best regards, The OBO Foundry Operations Committee.
Highlights
OBO Foundry Home Page now highlights ontologies that meet OBO Foundry principles
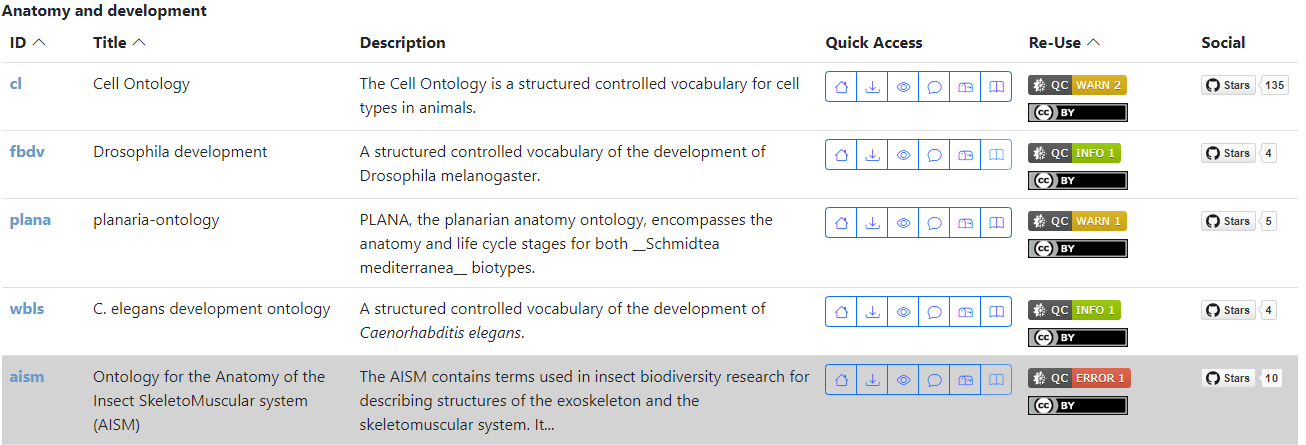
The OBO Foundry front page now visually promotes ontologies that pass the basic metadata requirements implemented by the OBO Dashboard (described below). Ontologies with no dashboard errors are listed first in their section; those with errors are at the end, with gray-shaded backgrounds (as shown in the screenshot above). We hope that ontology developers will take this as an incentive to update their metadata records! Get help in the #dashboard channel in the OBO-community Slack workspace. The team is ready to support you!
OBO Foundry dashboard analysis
The OBO Foundry Dashboard is a tool that automatically checks ontologies in the OBO Foundry for adherence to the OBO Foundry principles). It provides:
- A report on each ontology’s compliance with the 20 principles (green means an ontology meets the principle; pink means it does not; yellow means there was a warning). An ontology is considered to “pass” the dashboard if it has no errors.
- Overall statistics about the ontologies in the OBO Foundry, including the breakdown of how many ontologies currently pass the checks broken down by OBO principle (see Figure below).
The dashboard aims to inform both users and developers, allowing users to quickly assess an ontology’s quality and helping developers of the ontologies identify areas for improvement. The OBO Foundry announced last year that passing the dashboard would become mandatory for all ontologies on January 1, 2024, and that non-compliant ontologies would be listed at the bottom of their respective groups to encourage greater quality control.
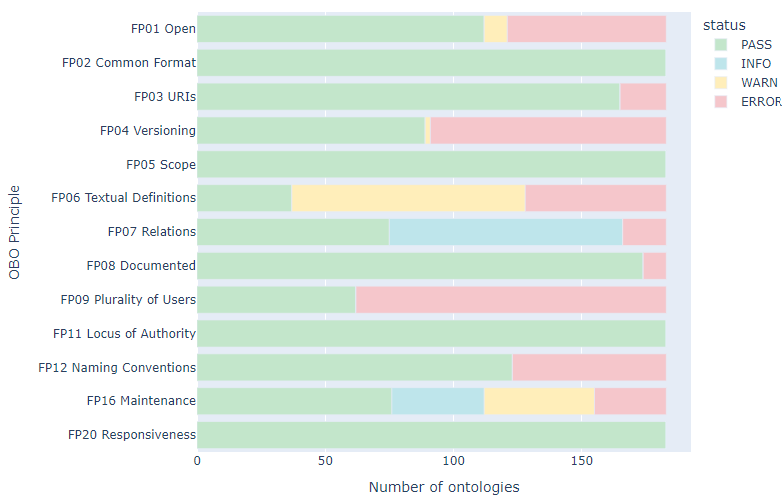
Above: The OBO dashboard showing the proportion of ontologies in the OBO Foundry that pass each active principle.
Decisions Made and Important Updates
- The committee proposed enforcing uppercase-only ID space prefixes for new submissions. Proponents argue that this change simplifies downstream tool processing and enhances consistency, while opponents highlight potential impacts on readability and issues with existing mixed-case prefixes. The vote ended 10 to 5 in favor of enforcing upper cases prefixes moving forward. This does not affect any existing ontologies with mixed-case prefixes.
Ongoing Discussions
Here, we list some of the discussions happening around the OBO-sphere:
- The recent meeting of the OBO Operations Committee (OBO Ops) addressed several important topics. A key requirement was emphasized: all OBO Ops members must belong to at least one working group—Editorial, Technical, or Outreach. The potential reorganization of these working groups was discussed to enhance efficiency and collaboration. Members of the OBO Operations team will be asked to reassign themselves to specific working groups, Additionally, the necessity of ensuring that all OBO Ops have clearly assigned tasks was highlighted, aiming to enhance accountability and productivity across the board. A discussion on the appropriate terminology for former members who continue to contribute is in progress to appropriately recognize their ongoing efforts. Additionally, the committee is addressing the issue of incentivizing active participation in working groups, aiming to foster greater engagement and contribution from all members.
- New ontology request issues with no updates from the submitter for more than 2 months will be closed with a standardized message. They can be re-opened by the submitter at any time they are ready to provide updates.
Ontologies
New ontologies accepted in the OBO Foundry Ontology Library \
- SLSO (Space Life Sciences Ontology) (SLSO) is an application ontology designed to support the NASA’s Life Sciences Data Archive and other systems that store space life science research data. The purpose of the ontology is to define and organize the specialized concepts and terminologies used in space life sciences research, which often involves expensive and specialized equipment, procedures, and references to space environments. The SLSO was created using the Ontology Development Kit (ODK) and incorporates concepts from various OBO Foundry ontologies. This allows for the indexing and integration of space biology investigation data with other scientific data related to space environments. The ontology was reviewed by Jie Zheng (see our Spotlight on members below) with active discussion with the maintainer of the ontology.
- AFPO (African Population Ontology) is an ontology that enables the annotation of African individuals and combines knowledge accumulated about existing populations with their genetic fingerprints in a standardized format. AfPO can be employed to comprehensively classify African study participants in prospective research studies. It can also be used to classify past study participants by mapping them using a language or ethnicity identifier or synonyms. The ontology was reviewed by Lynn Schriml with active discussion with the maintainer of the ontology.
New ontologies currently under review
- The Biomarker Ontology (BMO) is a comprehensive knowledge representation involving a variety of fields of medical and biological aspects for improving biomarker identification tasks, as well as a supportive integratable tool for abundant AI techniques, such as Machine Learning (ML) and Large Learning Model (LLM). It is being reviewed by Deepak Unni.
- The Physiologically-Based Pharmacokinetics (PBPK) ontology is a comprehensive framework designed to systematically capture and represent the intricate relationships and behaviors of pharmacokinetic processes within living organisms. It is being reviewed by Alex Diehl.
Spotlight on well-established OBO ontologies
In this issue, we continue our ontology spotlight series, highlighting two other ontologies from the OBO Foundry family, Chemical Entities of Biological Interest (ChEBI) and Vaccine Ontology (VO)
- Chemical Entities of Biological Interest (ChEBI) is a freely available, comprehensive database and ontology focused on small chemical compounds of biological relevance. It covers a wide range of molecular entities, including atoms, molecules, ions, and complexes, and provides detailed information on each entity, such as structure, formula, mass, charge, and names. ChEBI uses an ontological structure to organize entities and establish relationships, employing a controlled vocabulary for consistency. ChEBI is semantically integrated with over 100 biomedical ontologies, thus providing powerful capabilities for data integration, hypothesis generation, and reasoning. ChEBI also provides cross-reference links to several other databases and supports interoperability, making it widely used in various fields like cheminformatics, metabolomics, drug discovery, and systems biology. As of June 2024, ChEBI contains 61,325 fully curated entries, establishing it as the leading chemical ontology for life science applications and Semantic Web data annotation. ChEBI can be accessed online and downloaded for research purposes.
- As an OBO library ontology, the Vaccine Ontology (VO) is a community-based biomedical ontology in the domain of vaccine and vaccination. VO systematically standardizes and integrates various vaccines, vaccine components, vaccine mechanisms, vaccine data types, leading to the enhanced computer-assisted reasoning. Dr. Oliver He at the University of Michigan in 2007 initiated the VO development by collaborating with many OBO ontology developers including Drs. Barry Smith, Lindsay Cowell, Melanie Courtot, Bjoern Peters, Alan Ruttenberg, Chris Mungall, Richard Scheuermann, et al. The VO was first presented in the first International Conference of Biomedical Ontology (ICBO), Buffalo, NY, in July 2009 (Ref). Dr. Asiyah Yu Lin later joined He laboratory as a postdoctoral fellow and worked on the VO development during 2010-2014 (Ref). Compared to the 2009-06-08 version of VO that had 2,007 terms, the current VO version as of 2024-06-26 has a total of 14,120 terms, indicating its fast growth. In addition to modeling of vaccines against infectious disease vaccines, the VO has also been expanded to cover non-infectious disease vaccines, including the recently reported cancer vaccines (Ref). The cancer vaccine ontology modeling effort was led by Dr. Jie Zheng who has joined the University of Michigan since 2023 and has been working on this VO project (Jie is featured in our Spotlight on Members below). The VO and VO-related research has been funded by a NIH U24 grant (U24AI171008). In addition, VO has been widely used in many different projects, such as the ImmPort project (https://www.immport.org/), Human Immunology Project Consortium (HIPC) project (https://immunespace.org/), and the Vaccine Adjuvant Compendium (VAC, https://vac.niaid.nih.gov/).
Members and Volunteers
The OBO Foundry is honored to highlight two members who are making valuable contributions to our community:
Jie Zheng
###

Jie Zheng is a software developer and bioinformatician at the University of Michigan, Ann Arbor. She earned her Ph.D. in Biology from Queen’s University (Kingston, Canada) in 2009 and conducted postdoctoral research at the University of Pennsylvania under Dr. Chris Stoeckert, a pioneer in the field of biological ontologies. In 2014, Jie joined the OBO Foundry community and took part in its Operations Committee. She initiated and coordinated the development of the OBO Metadata Ontology to harmonize ontology metadata. Jie has worked on various OBO Foundry ontologies, such as the Ontology of Biomedical Investigations, Occupation Ontology, and Vaccine Ontology. Additionally, she was involved in the development of web-based ontology tools, Ontoanimal (e.g., OntoBee, OntoRat, and Ontodog), in collaboration with Dr. Yongqun “Oliver” He’s team. During her decade-plus working at UPenn, Jie’s focus was primarily on developing ontologies and leveraging them to establish metadata standards, build graph-based knowledge bases, and integrate and analyze data across diverse resources. In 2023, Jie joined Dr. He’s laboratory at the University of Michigan, where she now applies her extensive knowledge of ontologies combined with machine learning techniques to vaccine research.
Damion Dooley

Damion Dooley is an Ontology Development Lead at the Centre for Infectious Disease and One Health (CIDGOH.ca), a 20+ member group of bioinformatics and ontology researchers at the Faculty of Health Sciences, Simon Fraser University, Canada. His ontology work began in 2013 to support rapid enteric pathogen whole genome sequencing and analysis. Damion led the creation of the FoodOn food ontology and the Joint Food Ontology Workgroup to facilitate foodborne disease, agriculture, nutrition and food science data sharing. FoodOn, adopting the many food description facets of LanguaL, a popular food composition thesaurus, is now being used and supported by a wide variety of research and inter-agency data sharing projects. He also participates in The Open Biological and Biomedical Ontologies Foundry operations committee, and in curation of the Ontology for Biomedical Investigations (OBI). He looks forward to a future of FAIR, harmonized data that reduces barriers to discovery and insight in a rapidly changing world.
New Members of the OBO Operations Committee
In this issue we introduce our new member of the OBO Operations Committee. Welcome aboard, Jane Lomax!
Jane Lomax

Dr. Jane Lomax is currently the Head of Ontologies at SciBite, a semantic technology company. In this role, she oversees the development of ontological solutions for clients across various industries, including pharmaceutical R&D, healthcare and consumer goods. She is an advocate for the use of open ontologies and FAIR data principles in industry.
Jane has over 20 years of experience in research, development, and application of ontologies, with former roles with the Gene Ontology and WormBase at EMBL-EBI and the Wellcome Sanger Institute. Jane is an active participant in several public endeavors, she has served on the executive committee of the International Society of Biocuration (ISB), the ELIXIR GCBR Review Committee and presently holds a position on the Pistoia Alliance board.
Jane is a big advocate of the use of public ontologies in industry. At SciBite she supports customers to make best use of them, through both SciBite’s software and services. Jane joined the OBO Operations committee because she feels like there is much to be gained by establishing links between the commercial world and the OBO Foundry, and hopes, by taking up this position, to help facilitate that.
Spotlight on Research in the OBO community
The Artificial Intelligence Ontology: LLM-assisted construction of AI concept hierarchies
The Artificial Intelligence Ontology (AIO) is a systematization of artificial intelligence (AI) concepts, methodologies, and their interrelations. Developed via manual curation, with the additional assistance of large language models (LLMs), AIO aims to address the rapidly evolving landscape of AI by providing a comprehensive framework that encompasses both technical and ethical aspects of AI technologies. The primary audience for AIO includes AI researchers, developers, and educators seeking standardized terminology and concepts within the AI domain. The ontology is structured around six top-level branches: Networks, Layers, Functions, LLMs, Preprocessing, and Bias, each designed to support the modular composition of AI methods and facilitate a deeper understanding of deep learning architectures and ethical considerations in AI.
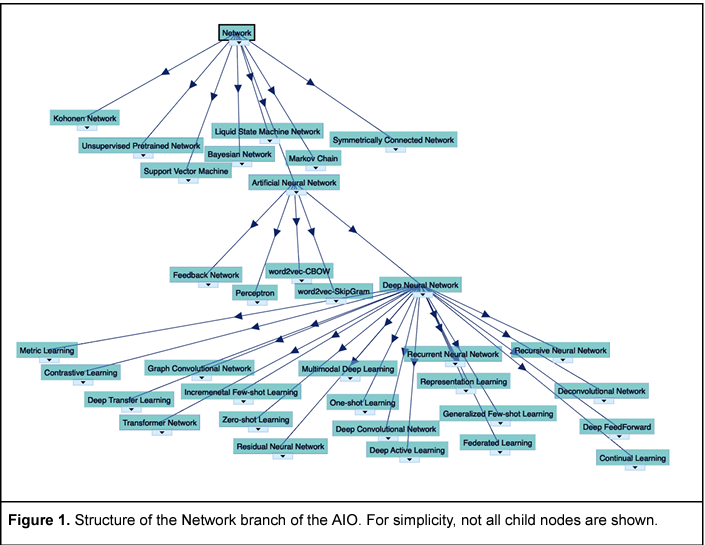
(Figure from Joachimiak M p., Miller MA, Caufield JH, Ly R, Harris NL, Tritt A, Mungall CJ, Bouchard KE. The Artificial Intelligence Ontology: LLM-assisted construction of AI concept hierarchies. 2024 Apr 3; http://arxiv.org/abs/2404.03044)
Empowering standardization of cancer vaccines through ontology: enhanced modeling and data analysis
This work by Zheng et al (Jie Zheng was also featured above in the OBO Member Spotlight) is about the development of an ontological model for cancer vaccines using the Vaccine Ontology (VO) to facilitate standardization, integration, and analysis of heterogeneous cancer vaccine data. The authors used various ontologies and tools from the Open Biomedical Ontologies (OBO) Foundry to develop a comprehensive ontological model for cancer vaccines within the Vaccine Ontology (VO). They followed OBO Foundry principles by reusing terms from existing ontologies such as the Human Disease Ontology (DOID), NCBITaxon, and the Ontology for Biomedical Investigations (OBI). Tools like ROBOT and the Ontology Development Kit (ODK) were used to extract relevant terms, automate ontology workflows, and ensure adherence to OBO guidelines. With this approach, the authors created a Cancer Vaccine Ontology (CVO) that standardizes diverse cancer vaccine data, supports ontology-based analysis of vaccine attributes and clinical trials, and facilitates data integration and knowledge discovery.
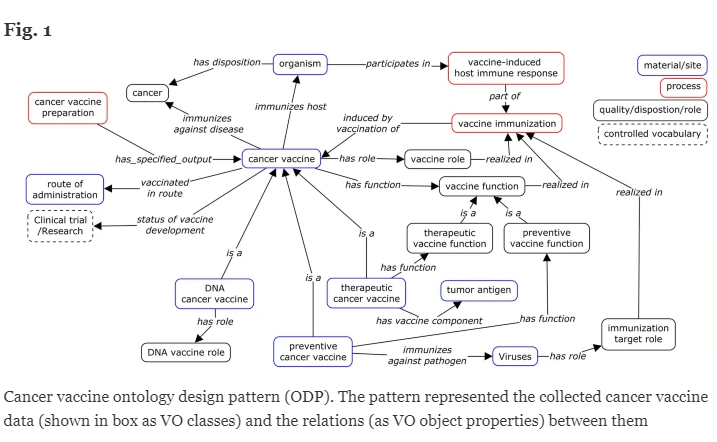
(Figure from Jie Zheng, Xingxian Li, Anna Maria Masci, Hayleigh Kahn, Anthony Huffman, Eliyas Asfaw, Yuanyi Pan, Jinjing Guo, Virginia He, Justin Song, Andrey I. Seleznev, Asiyah Yu Lin & Yongqun He. 2024 June 19 https://jbiomedsem.biomedcentral.com/articles/10.1186/s13326-024-00312-3
Spotlight on OBO Principles
In this issue, we continue with Principle 4 of the OBO Foundry’s series of principles.
OBO Foundry Principle 4: Versioning
Principle 4 specifies the requirements for versioning. The ontology provider must have well-documented procedures in place for versioning the ontology. Each official release of the ontology must be clearly marked with a unique version identifier, which should be in one of two formats: either a date format following the “YYYY-MM-DD” pattern or a numbering system such as semantic versioning (e.g., “NN.n”).
Every released version must have a unique IRI version that resolves to the specific ontology artifact associated with that version. The IRI must contain the same version identifier as the artifact itself to ensure consistency. It is recommended that version IRIs use dated PURLs to ensure long-term accessibility.
Once an official version of the ontology is released, the content of the released files must not be altered. If any bugs or issues are discovered in a released version, they should not be fixed by modifying the existing files. Instead, the fixes should be incorporated into a new official release with its own unique version identifier and corresponding files.
Additionally, the ontology should include an owl:versionInfo statement that matches the version identifier used in the IRI and the artifact.
Fixing common problems
The automatic check that is performed as part of the OBO Dashboard is implemented here. Roughly half of the ontologies in the OBO Foundry currently pass this check. While there are some ontologies that do not contain a correctly formatted version IRI, the most frequent problem is that an existing version IRI does not resolve correctly. A typical solution to this is to ensure that an appropriate rule (see, for example, PATO) for redirecting from GitHub is defined, and then use GitHub releases as a way to create “tags”. For example, when the March 2024 release of PATO was created (https://github.com/pato-ontology/pato/releases/tag/v2024-03-28), it was created using the “tag” v2024-03-28 (note the leading “v”). This tag works very similar to a branch, and you can actually browse the GitHub repository for that tag (https://github.com/pato-ontology/pato/tree/v2024-03-28). In summary, for 90% of the problems:
- Make sure you have a correctly formatted version IRI that is updated with ever release
- Every time you create a release, you also create a GitHub release
- The purl config redirects the version IRI to the GitHub tag (as shown in the PATO example above)
Spotlight on Tools: Recent updates
Special announcement to the entire OBO community since we’ve had not one, not two, but three software releases of interest in the past few days:
ROBOT 1.9.6: In the latest update, several additions and changes have been made. The duplicate exact synonym report query has been updated to be case-insensitive and to ignore synonyms annotated as abbreviation or acronym synonym types (#1179). The –drop-axiom-annotations option has been extended to support value-specific removal of axiom annotations (#1193). The obographs have been updated to version 0.3.1, and the OWL API has been upgraded to version 4.5.29, which includes a major update to the OBO Format, now supporting IDSPACE declarations for non-OBO Foundry prefixes. The Elk reasoner has also been updated to version 0.6.0.
ODK 1.5.1: In the ODK toolbox, ROBOT has been updated to version 1.9.6 (see above). It also includes an updated version of the Elk reasoner and long-awaited prefix map support for the OBO format.
Protégé 5.6.4: For better interoperability, this minor release uses the same latest OWLAPI version as the one used in ROBOT 1.9.6, and the same latest ELK reasoner as well. It fixes a few bugs, makes the support for ID range policy files more flexible, and adds a new setting to configure which explanation plugin(s) should be used.
Spotlight on Tools: Ontology Browsers
Here and in upcoming issues of this newsletter, we would like to highlight some key browsers in the ontology community: the Ontology Lookup Service (OLS), Ontobee, and BioPortal. These browsers are essential for navigating and utilizing the support of ontological data available. We spotlight OLS in this issue; watch for spotlights on Ontobee and BioPortal in future issues.
Ontology Lookup Service (OLS)
The Ontology Lookup Service (OLS) is a critical tool for the OBO Foundry community, providing a comprehensive and user-friendly interface for accessing and searching a wide array of OBO ontologies. OLS simplifies the process of exploring and integrating ontological data by offering a centralized platform where users can browse, query, and visualize terms across multiple OBO Foundry ontologies. With its API and advanced search capabilities, OLS enables researchers and developers to efficiently retrieve relevant information and incorporate ontological knowledge into their applications. OLS continues to support the OBO Foundry community in managing and utilizing vast amounts of structured data effectively.
Events
- ICBO 2024 in the Netherlands
- The OBO Foundry Tutorial at ICBO will focus on collaborative workflows and leveraging freely available online training resources. Attendees will learn strategies for maintaining community-based ontologies in a distributed development environment. The goal of the tutorial is to provide insights into the latest updates and best practices in ontology development. Here are some key topics that will be covered:
- Managing ontologies built and maintained by volunteers and dispersed teams
- Utilizing GitHub for issue tracking, documentation, discussions, automation pipelines, and highlighting social workflows
- Adhering to open science principles
The tutorial will have a “show and tell’’ format to encourage real-time participation. Attendees are encouraged to install the following software in advance: GitHub Desktop, Protégé, and ROBOT. The tutorial is organized by Nico Matentzoglu from Semanticly and Nicole Vasilevsky from the Critical Path Institute.
- The OBO Foundry Tutorial at ICBO will focus on collaborative workflows and leveraging freely available online training resources. Attendees will learn strategies for maintaining community-based ontologies in a distributed development environment. The goal of the tutorial is to provide insights into the latest updates and best practices in ontology development. Here are some key topics that will be covered:
- COB Workshop: Purpose: To advance the development of COB.
- The COB Workshop will be held at the La Jolla Institute for Immunology (9420 Athena Cir, La Jolla, CA 92037) on September 23-24, 2024. Register for the workshop at https://forms.gle/QZetUv2VhPhM3Rqq6.
- Both in-person and virtual attendees are required to complete the registration form in order to receive additional information.
- OBO Academy: Monarch Seminar Series (ongoing)
- The OBO Academy: Monarch Seminar Series has been moved to a monthly cycle until further notice. The next seminar on 2024/07/23 will be a follow up session on the ICBO tutorial above, see schedule below. After that, OBO Academy will go into a summer break, and we will resume 2024-09-17 (topic tbd).
- Schedule: https://oboacademy.github.io/obook/courses/monarch-obo-training/
- You can vote for future training topics here: https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests
Recent OBO Academy seminars
- Introduction to the Uberon Anatomy ontology (Damien Goutte-Gattat (FlyBase) and Nico Matentzoglu)
- An Introduction to Synonyms in OBO Ontologies (Nicole Vasilevsky, C-Path)
- AI-assisted ontology editing workflows 2: Validation (Chris Mungall, LBNL)
- Phenotype data and the role of ontologies (James McLaughlin (EBML-EBI) and Nico Matentzoglu)
Ways to be part of the OBO Foundry community
There are many ways to be part of the OBO Foundry community, beyond using our website to find ontologies of interest. For example:
- Join the obo-discuss mailing list (low-traffic, no spam)
- If you are interested in the technical aspects, you can also join the obo-tools mailing list
- Join the conversation in our Slack workspace (when you join the obo-discuss mailing list, the welcome message has an invitation link for the Slack workspace).
- Use our public issue tracker to report a problem you discovered on obofoundry.org or request a new feature
- Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be added to the issue tracker for the specific ontology
- Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of our code is publicly readable, you may be able to pinpoint where a bug fix needs to go.
- Request that your ontology be considered for inclusion in the Foundry
- If you want to take your contributions to OBO Foundry to the next level, you can nominate yourself to be considered for the OBO Operations Committee. There are various roles in which you can contribute, including assisting with the production of this newsletter