OBO Foundry Newsletter Issue 7
Happy New Year from the OBO Foundry Community!
In this edition, we highlight two of our dedicated community members and two OBO Foundry ontologies. You will also find updates on recent workshops, the latest tools to improve ontology development, and the progress of current projects.
As always, we encourage you to share your ideas, feedback, and suggestions with the OBO Foundry Operations Committee.
Highlights
OBO Dashboard updated
The OBO Foundry Dashboard tracks the progress of ontologies in meeting OBO Foundry principles, such as good metadata and interoperability. Ontologies that meet these standards are highlighted on the OBO Foundry homepage at https://obofoundry.org. We encourage all ontology developers to use the dashboard to check their ontologies and improve them. You can access the dashboard (last updated 2025-01-16) here: OBO Dashboard.
Ongoing Discussions
Editorial Working Group
The EWG focuses on refining the principles and guidelines that ensure consistency and quality across OBO ontologies.
- Principle 19, “Term Stability”: Revisions to Principle 19, focusing on how to handle obsolete terms, dominated the EWG’s recent discussions. The EWG decided to implement a check for the owl:deprecated property on classes containing “obsolete” or similar variations in their name or definition. They are also exploring methods, such as analyzing GitHub issue complaints, to assess term stability more comprehensively. The OBO community is welcome to comment on the proposed revisions to this principle.
- Principle 7, “Relations”: Discussions around Principle 7 aimed to provide clearer guidance on when relations should be added to the Relation Ontology (RO) or kept within a specific ontology. This is a recurring challenge, and the EWG proposed conditions focusing on a relation’s domain and range and its potential for broader use. A significant concern is preventing “relation striping,” where related terms end up defined across multiple ontologies. The proposed guidance is:
- If both the domain and range of a proposed relation belong to the same ontology, it is acceptable to keep the relation within that ontology. However, it should still have a parent term in RO if one exists.
- If only the domain of a relation belongs to the same ontology, the decision to include it in RO depends on whether the relation is invertible. Invertible relations, where the subject and object can be swapped, raise additional considerations.
- Relations with a domain outside the defining ontology should be added to RO.
- Relations considered generally usable (applicable in other ontologies) should also be added to RO.
- When in doubt, add the relation to RO. This principle encourages a preference for including relations in RO whenever possible. The EWG’s recommendations emphasize the importance of RO as the central repository for relations, promoting interoperability and consistency across OBO ontologies. However, several challenges and open questions remain:
- RO’s Comprehensiveness: RO might not yet be comprehensive enough to cover all possible relation types. What happens when a suitable RO parent term doesn’t exist for a relation that meets the criteria for remaining within a specific ontology?
- “Relation Striping”: This refers to the problem of related relations being defined across different ontologies, hindering clarity and findability. The EWG is considering how to prevent this, potentially by retroactively moving related relations into RO.
- COB’s Use of RO Relations: The relationship between COB and RO, specifically which relations COB should hold, is another ongoing discussion.
- The EWG is actively working on addressing these challenges and refining Principle 7 to provide clearer and more actionable guidance for ontology developers. This will improve the consistency and interoperability of relations across OBO ontologies.
- Naming Conventions: The EWG reported persistent issues with inconsistencies in naming conventions across ontologies. This includes variations in capitalization, the use of camel case and underscores, and the application of title case. Though each of these are considered violations of Principle 12 “Naming Conventions”, there are numerous cases where such are warranted, such as capitalized genus names in taxonomies. Acknowledging the need for exceptions in certain cases, the EWG is working towards establishing mechanisms to distinguish between warranted exceptions and true violations.
Discussions are ongoing, with the committee actively seeking feedback (which you can provide by commenting on Principles 7 and 12) and working towards revised versions of these principles.
Ontologies
New ontologies currently under review
- PRIDE (Proteomics Identification Ontology): The PRIDE (Proteomics Identification) Ontology provides a structured framework for describing and sharing proteomics data from mass spectrometry experiments, organizing information in the PRoteomics IDEntifications Database. The ontology includes terms for protein identifications, peptide sequences, experimental conditions, and modifications, enabling researchers to consistently annotate their data across different studies. Join the discussion.
- Membrane Ontology (MEMON): The Membrane Ontology creates a structured vocabulary for annotating biological data about cell membranes and their experimental and computational models, allowing detailed membrane descriptions and connections between datasets. The terms cover different biological membrane models used in research, along with their component descriptions, and will expand to link models with the types of biological membranes they approximate. Join the discussion.
Spotlight on well-established OBO ontologies
In this issue, we continue our ontology spotlight series, highlighting two ontologies from the OBO Foundry family, the FlyBase Controlled Vocabulary (FBcv) and the Mammalian Phenotype Ontology (MP).
- The Mammalian Phenotype Ontology (MP) provides a structured vocabulary for curation and retrieval of mammalian phenotypes. Originally developed for use by Mouse Genome Informatics (MGI) and Rat Genome Database (RGD) over 24 years ago, the MP currently consists of more than 14,200 pre-composed phenotype terms organized across 28 broad categories. These terms have been used to curate phenotypes for over 74,800 mouse models in MGI. New releases are published on a monthly basis and are available from the MGI download page or the MP GitHub repository. New terms are added based on user requests and as part of systematic reviews and revisions. Recent updates have focused on expanding coverage for models of pediatric diseases that are part of the Gabriella Miller Kids First initiative (including idiopathic scoliosis, cleft palate, and congenital heart disease), reproductive system terms with a focus on infertility models, and COVID-19 models. During revisions we work on enhancing the alignment between the MP and the Human Phenotype Ontology (HPO) both by improving consistency of logical definitions across the ontologies and by manual mapping of MP and HPO terms using the SSSOM format. The mapping files are available in the Mouse-Human Ontology Mapping Initiative GitHub repository. The MP has become an integral part in disease model identification and disease gene prediction and the ongoing work will enhance the utility of the MP for researchers, clinicians, and curators. Questions and suggestions should be directed to the MP GitHub issue tracker (https://github.com/mgijax/mammalian-phenotype-ontology/issues).
- The FlyBase Controlled Vocabulary (FBcv) has existed as an ontology since at least 2007 and was intended to provide controlled terms for describing Drosophila genetic entities and associated data. Alleles in FlyBase have detailed, standardised annotations, with information about encoded tools (such as GFP) and types of functionality (e.g. conditional loss of function) curated using FBcv. Phenotype curation uses the Drosophila Phenotype Ontology, which is incorporated into FBcv (as well as being available as a standalone file) plus qualifier terms (such as ‘somatic clone’), which are also found in FBcv. There are also terms to describe publications (e.g. ‘paper’) and datasets, including methodologies and sample attributes. FBcv has been on GitHub since 2015 and using the Ontology Development Kit since 2019. Most term additions over the past year have been new types of genetic tools, reflecting the diverse manipulations possible in this model organism.
Members and Volunteers
The OBO Foundry is honored to highlight two valued community members.
Nicole Vasilevsky

Nicole Vasilevsky is the Associate Director of Data Science at the Critical Path Institute (C-Path), which is based in Tucson, AZ (USA) but she works remotely with her cats from the coffee-loving city of Portland, OR (USA). Her journey into the world of ontology curation began in the lab of Melissa Haendel at Oregon Health & Science University, where she was initiated into ontology development through her work on the eagle-i Network. As one of the original members of the Monarch Initiative, she helped develop tools that utilize ontologies for disease diagnostics. She has contributed to various OBO ontologies, including the Human Phenotype Ontology (HPO) and Mondo Disease Ontology, where she served as the original curator and lead and remains an active contributor. After sharing fears about “breaking” ontologies with colleagues at the International Conference on Biomedical Ontology (ICBO), Nicole began contributing to the OBO tutorials at ICBO to train ontology developers. This experience led her to consult at C-Path, collaborating with colleagues to train team members in ontology development and laying the foundation for the OBO Academy. Since joining Ramona Walls’ team at C-Path in January 2023, she is currently focused on standardizing rare disease data through the Rare Disease Cures Accelerator - Data and Analytics Platform (RDCA-DAP). Nicole serves on the OBO Foundry Operations Committee and values the mentorship and collaborations she’s fostered in the community and hopes our work will enhance our understanding and treatment of rare diseases.
Chris Mungall

Dr. Chris Mungall is a Senior Scientist in the Environmental Genomics and Systems Biology Division at Lawrence Berkeley National Laboratory, where he heads the Biosystems Data Science department. Chris’s research involves the curation and integration of biological research data to elucidate biological mechanisms underpinning the health of humans and of the planet. He is a PI on both the Gene Ontology (GO) project and the Monarch Initiative.
Chris has a background in knowledge representation, and was one of the original founding members of the OBO Foundry. He has led or made major contributions to multiple ontologies such as the GO, Uberon, ENVO, Mondo, and RO. He has helped pioneer ontology-related standards such as LinkML, the Knowledge Graph Change Language (KGCL), and SSSOM, and his team has helped build an ecosystem of tools around OBO, including the Ontology Access Kit (OAK), and LLM-based tools including OntoGPT.
In 2017, Chris was the first person to be awarded the Exceptional Contributions to Biocuration Award by the International Society for Biocuration.
Spotlight on Research in the OBO community
A change language for ontologies and knowledge graphs
Ontologies and knowledge graphs evolve constantly as scientific knowledge grows. A new framework, KGCL (Knowledge Graph Change Language), offers a standardized way to track and implement these changes. KGCL uses clear commands like “rename X to Y” that both humans and computers can understand.
The system comes with practical tools: Ontobot processes changes through GitHub automatically, while a new BioPortal widget lets users suggest updates through a simple interface. These tools are already being used by Mondo, GO, ENVO, and Uberon. KGCL helps ontology teams work more efficiently while making it easier for the broader scientific community to contribute.
The development of KGCL was driven by community needs, in particular a 2023 workshop attended by many OBO ontology developers and curators. As ontologies continue to grow in importance for biological research, KGCL offers a timely solution for keeping these vital resources up to date.
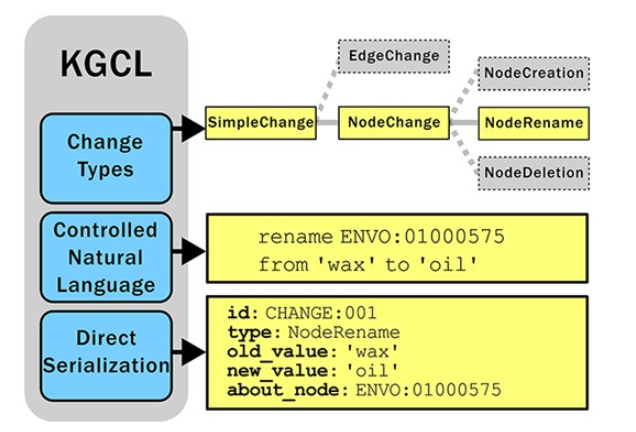
Figure from Hegde H, Vendetti J, Goutte-Gattat D, Caufield JH, Graybeal JB, Harris NL, Karam N, Kindermann C, Matentzoglu N, Overton JA, Musen MA, Mungall CJ. A change language for ontologies and knowledge graphs. 2025 Jan 22 https://doi.org/10.1093/database/baae133.
Whelk: An OWL EL+RL Reasoner Enabling New Use Cases
Many of the ontologies in OBO are large, with tens of thousands of classes, many of these classes interlinked via OWL axioms. Some of these ontologies are too large to use with most OWL reasoners. A major game changer for the OBO world was the release of the ELK reasoner, which was fast enough to reason over large ontologies like GO, CL, and Uberon. One reason ELK is so much faster than other reasoners is that it operates over a restricted subset or profile of the OWL language (formally called EL++). However, this tradeoff means that ELK can’t operate over all axiom types, including certain kinds of rule axioms found in the Relation Ontology.
Whelk is a new reasoner that allows for reasoning over both OWL EL, as well as the OWL RL profile, giving ontology editors additional expressivity when reasoning with instance graphs and rules. Developed in Scala with immutable functional data structures, Whelk excels in handling high-throughput queries and dynamic reasoning tasks, outperforming traditional reasoners like ELK in specific scenarios.
The reasoner supports complex class expressions and SWRL rules, enabling rapid instance reasoning, parallel query processing, and incremental updates without affecting prior ontology states. Applications include relation graph materialization, biomedical ontology reasoning, and enhanced query performance for large ontologies such as Uberon and the NCI Thesaurus.
Whelk demonstrates its utility in real-world scenarios, such as the Ubergraph knowledge graph pipeline, executing millions of DL queries efficiently, allowing for precomputation of different valid paths through the ontology.
Whelk is available as a Protege plugin, and can be easily dropped into existing ODK pipelines, or used in combination with ROBOT. Whelk has already been adopted as the default reasoner by the Cell Ontology (CL).
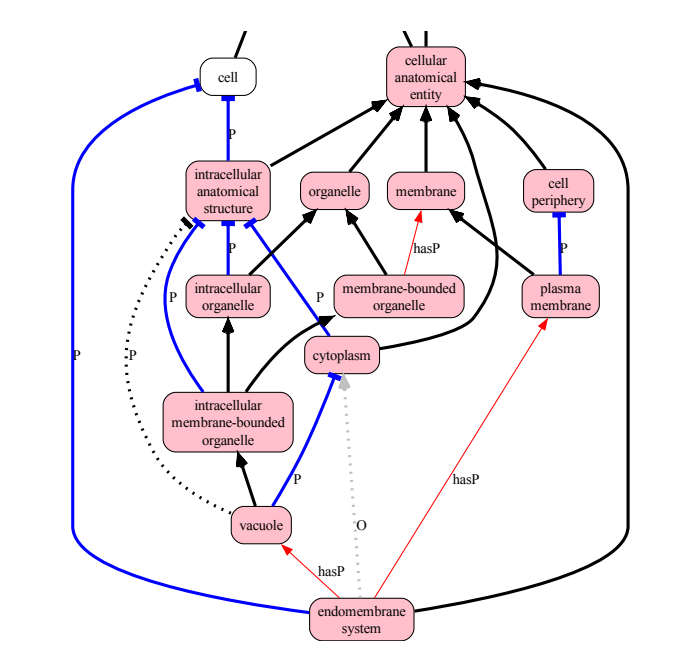
Figure from James P. Balhoff (University of North Carolina, Chapel Hill) and Christopher J. Mungall (Lawrence Berkeley National Laboratory). Whelk: An OWL EL+RL Reasoner Enabling New Use Cases. 2024 Dec 18; https://doi.org/10.4230/TGDK.2.2.7
A general strategy for generating expert-guided, simplified views of ontologies
This preprint introduces a strategy for creating expert-guided, simplified views of complex biomedical ontologies to support FAIR data practices. Caron et al. (2024) address challenges faced by researchers using detailed ontologies like Uberon and Cell Ontology (CL) by developing tools to validate user hierarchies and generate tailored ontology views.
Applied in projects like HuBMAP and Human Developmental Cell Atlas (HDCA) these workflows simplify ontology use while ensuring accuracy. Automated validation pipelines and visualization tools enhance alignment between user needs and ontology structures, fostering collaboration between researchers and curators.
The tools, available at https://github.com/hubmapconsortium, offer a practical approach to integrating ontology-driven data annotation and analysis.
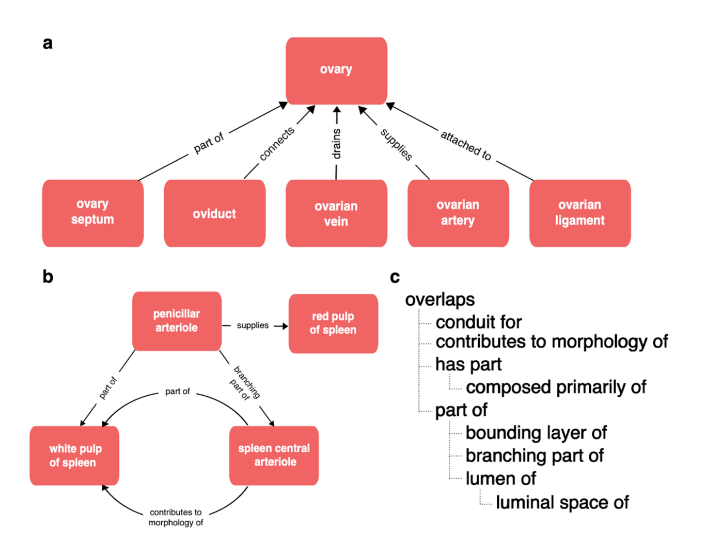
Figure from Anita R. Caron, Aleix Puig-Barbe, Ellen M. Quardokus, James P. Balhoff, Jasmine Belfiore, Nana-Jane Chipampe, Josef Hardi, Bruce W. Herr II, Huseyin Kir, Paola Roncaglia, Mark A. Musen, James A. McLaughlin, Katy Börner, and David Osumi-Sutherland. A General Strategy for Generating Expert-Guided, Simplified Views of Ontologies. 2024 Dec 18; https://doi.org/10.1101/2024.12.13.628309)
Spotlight on OBO Principles
Principle 6 of the OBO Foundry emphasizes the importance of textual definitions for ontology terms. A textual definition is a clear, human-readable explanation of what a term represents, helping researchers understand its meaning and use. These definitions should be unique, written in English, and ideally follow a structure like “a B that Cs,” where B is the broader category and C specifies the unique characteristics. Textual definitions should align with logical definitions (used by machines) to avoid confusion in classification or annotation.
The principle allows flexibility through alternative approaches - terms that resist strict definition can use elucidations, and definitions can be generated programmatically using Dead Simple Ontology Design Patterns (DOSDPs). For optimal clarity, usage examples and editorial notes should be kept separate from the core definition. Though instances like organizations or locations may have definitions, they aren’t required for these cases.
We recommend that anyone who writes ontology definitions reads this excellent blog post by Chris Mungall from 2019: https://douroucouli.wordpress.com/2019/07/08/ontotip-write-simple-concise-clear-operational-textual-definitions/.
While the OBO Dashboard only reports a warning when classes are not defined (for practical reasons), we want to remind the community that definitions are important to enable both humans and AI agents to understand and recognise the concept defined. Labels alone are often ambiguous. You are doing your human users and future AI assistants a big favour if you take writing human-readable definitions seriously.
Spotlight on Tools: Recent updates
Babelon for ontology translations
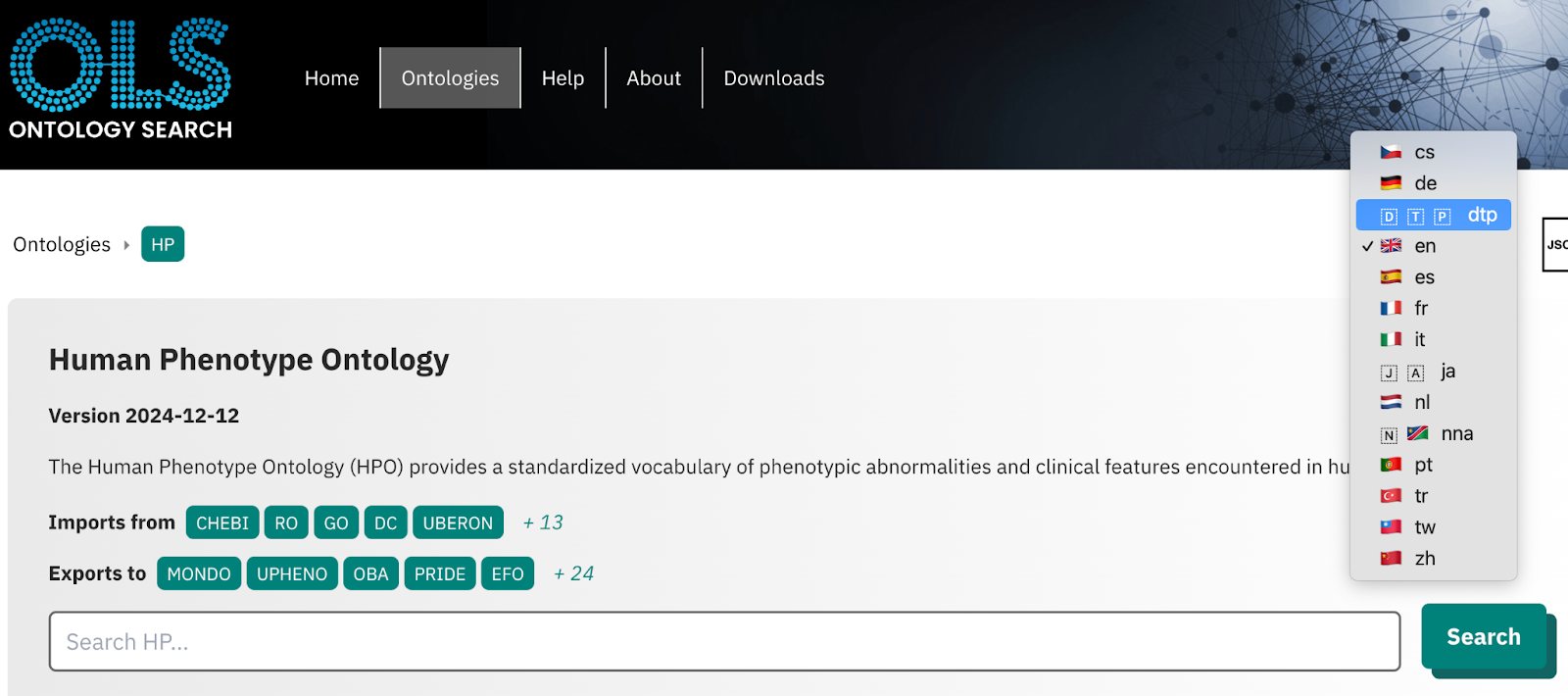
Babelon for ontology translations. The Babelon format is a simple spreadsheet-based format to curate and integrate ontology translations. It has been implemented by various widely used ontologies such as HPO, MP and Mondo. It can be used by ODK-managed ontology systems to publish “international editions” of an ontology. Those international editions can be leveraged by ontology browsers such as OLS to display the ontology in various languages; see below. The Babelon toolkit supports operations such as simple, AI-assisted translation of the entire ontology. Some example Babelon files can be found here. More information about Babelon workflows can be found in this presentation.
ROBOT-Template Helper
The ROBOT-Template Helper GPT is a tool designed by Chris Mungall to assist researchers in creating ontology templates for use with ROBOT, a set of tools for building ontologies that is commonly employed in the OBO (Open Biological and Biomedical Ontologies) community. It helps users generate structured CSV files that define ontology classes, relationships, and metadata in a format compatible with ROBOT’s template system. The tool supports adding annotations, logical axioms, and mappings to ontology terms, ensuring alignment with best practices for ontology development. By simplifying template creation, it enables users to efficiently build and manage domain-specific ontologies. For more information please check AI Guided Ontology Curation Workflows and the ROBOT Template GPT helper
Spotlight on Newly Approved Grants
Developing an Ontology for Dental Care-Related Fear and Anxiety (ODFA): Toward an Understanding of Problems in Dental Care Utilization
Principal Investigators: Bill Duncan, Daniel McNeil
NIH Reporter: https://reporter.nih.gov/search/LGKYr0JMlkauBjcllqzKaw/project-details/10939840
Dental care-related fear, anxiety, and/or phobia (hereafter, DFA) traditionally refers to the emotional, behavioral, and/or physical responses that may occur when thinking about or engaging in dental care. There is ample evidence in the scientific literature that DFA is associated with greater prevalence of oral conditions and diseases, including dental caries, tooth loss, and periodontal disease, leads to avoidance of both preventive and restorative dental care, and impairs oral health-related quality of life.
Despite the impact of DFA on oral health, the scientific literature is replete with terminology that refers only to dental fear or only to dental anxiety, yet in the broader psychological literature, fear and anxiety are known to be separate constructs with unique manifestations. This lack of consistency and interoperability between the medical and dental care communities in defining and classifying such phenomena has contributed to the current stalemate in scientific progress as it relates to understanding the etiology and implications of such phenomena in the dental care context. Without a consensus on the definition, types, scope, and etiology of DFA, the associated individual, clinical, and population impacts, and viable strategies for intervening to mitigate or manage such impacts the impact of DFA cannot be adequately studied. To address these terminological shortcomings, we have created the Ontology of Dental care-related Fear, and Anxiety, and/or Phobia (ODFA). By more precisely representing the types of fear and/or anxiety experienced by individuals, the ODFA’s concepts and relations facilitate the development of tools and resources capable of enhancing our understanding of the individual, clinical, and population impacts of dental care-related fear and anxiety. When applied to study data, the ODFA enables the integration and interoperability of data from multiple studies, which, in turn, provides a means to perform rigorous analysis on larger datasets in order to gain an in-depth understanding of the phenomenon and treatments for DFA.
Spotlight on Tools: Ontology Browsers
Popular ontology browsers include the Ontology Lookup Service (OLS), Ontobee, and BioPortal. We spotlight OntoBee in this issue.
Ontobee
Ontobee is the default resolver for many OBO ontologies and an essential tool for exploring them. Every ontology page on the OBO website includes a link to view that ontology in Ontobee. A major strength of Ontobee is its ability to display complex OWL axioms for ontology terms, which can also be retrieved programmatically. This makes it a valuable resource for ontology term dereferencing, linkage, and integration.
Ontobee is a collaborative project involving different parties. The source code of the Ontobee software is developed and maintained by the He Group. Ontobee performs weekly updates, automatically retrieving ontology sources and metadata from the OBO Foundry. Ontobee is part of a suite of tools (including Onto-Animals) that offer complementary features to support ontology exploration and usage, which will be highlighted in future newsletters.
For Ontobee software questions and feature requests, contact the Ontobee-discuss group (ontobee-discuss@googlegroups.com) or submit issues on GitHub (https://github.com/ontoden/ontobee/issues). Principal Contact: Dr. Yongqun “Oliver” He, University of Michigan Medical School, Email: yongqunhe@gmail.com
For further details, see:
Edison Ong, Zuoshuang Xiang, Bin Zhao, Yue Liu, Yu Lin, Jie Zheng, Chris Mungall, Mélanie Courtot, Alan Ruttenberg, Yongqun He, Ontobee: A linked ontology data server to support ontology term dereferencing, linkage, query and integration, Nucleic Acids Research, Volume 45, Issue D1, January 2017, Pages D347–D352, https://doi.org/10.1093/nar/gkw918.
Events and News
-
ChEBI workshop. The ChEBI workshop was held on 19th November 2024 at the European Bioinformatics Institute, Hinxton, Cambridgeshire, United Kingdom. ChEBI (Chemical Entities of Biological Interest) is a freely available database and ontology focused on small molecular entities. It provides a curated repository of chemical information that supports the life sciences research community.
The 1-day workshop was conducted as a hybrid event, with 24 in-person and 12 remote participants. Its primary aim was to bring together stakeholders from major bioinformatics resources that rely on ChEBI, provide updates on ChEBI’s redevelopment, receive feedback, and solicit further input. Representatives from various bioinformatics resources, including GO, UniProt, Rhea, MetaboLights, OLS, EuropePMC, PubChem, Reactome, IEDB, GlyGen, and LIPID MAPS, were present. Eloy Felix provided an overview of ChEBI’s history, technical landscape, and future goals. Muhammad Arsalan presented updates on ChEBI’s new web interface, advanced search functionality, and manual/automatic cross-references. Adnan Malik discussed the new submissions portal, while Carlos Moreno explained ChEBI’s ontology generation process and download formats. Juan Mosquera covered the new REST web services and the implementation of some new endpoints. The meeting concluded with Andrew Leach initiating discussions about ChEBI’s operating model and future funding. Additionally, five representatives from different resources gave talks introducing their resources, explaining how ChEBI is used within them, and suggesting potential improvements. Ozgur Yurekten presented MetaboLights, Kristian Axelsen presented Rhea, Peter D’Eustachio presented Reactome, Chris Mungall presented GO’s needs for a simpler version of CHEBI, and Randi Vita presented IEDB. The day was productive, marked by active discussions and constructive feedback. Many participants appreciated the idea of further developing ChEBI through community efforts, akin to the GO consortium model—opening up curation of the ontology part of CHEBI to a set of trusted partners. Some of the workshop slides can be found here: https://drive.google.com/drive/folders/1XMlzZFXXm7styUhzOBgpWchiwLutQj4f.
-
The 18th Annual International Biocuration Conference:
The 18th Annual International Biocuration Conference will take place from April 5 to 9, 2025, in Kansas City, Missouri, hosted by the Stowers Institute for Medical Research. This conference serves as a platform for professionals involved in the curation, development, and utilization of clinical and life sciences data to share their work and collaborate. Keynote speakers include experts from institutions such as Ohio State University, the University of Minnesota, the Allen Institute, the University of Arizona, EMBL-European Bioinformatics Institute, and the University of Southern California. Registration is open until February 28, 2025, or until capacity is reached. The event is organized in collaboration with the International Society for Biocuration (ISB).
- OBO Academy: Monarch Seminar Series (ongoing)
- Schedule: https://oboacademy.github.io/obook/courses/monarch-obo-training/
- You can vote for future training topics here: https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests
Ways to be part of the OBO Foundry community
There are many ways to be part of the OBO Foundry community, beyond using our website to find ontologies of interest. For example:
- Join the obo-discuss mailing list
- If you are interested in the technical aspects, you can also join the obo-tools mailing list
- Join the conversation in our Slack workspace.
- OBO Foundry RSS feed at https://obofoundry.org/feed.xml
- Use our public issue tracker to report a problem you discovered on obofoundry.org or request a new feature
- Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be added to the issue tracker for the specific ontology
- Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of our code is publicly readable, you may be able to pinpoint where a bug fix needs to go.
- Request that your ontology be considered for inclusion in the Foundry
- If you want to take your contributions to OBO Foundry to the next level, you can nominate yourself to be considered for the OBO Operations Committee. There are many ways in which you can contribute, including assisting with the production of this newsletter.